4 Study overview
4.1 Learning objectives
- Explain why spatial transcriptomics is used to study immune cell organisation
in IBD
- Describe how the data set is structured with healthy and disease cohorts
- Evaluate how replicates and balanced sampling improves the reliability of spatial transcriptomic studies
4.2 Background
This workshop will use CosMx Spatial Molecular Imager (SMI) data from the paper Macrophage and neutrophil heterogeneity at single-cell spatial resolution in human inflammatory bowel disease (Garrido-Trigo et al. 2023)1.
This data set was chosen because the authors have made their raw and annotated data available, along with the corresponding analysis code and a browsable data interface.
The study provides a clear biological contrast between health and disease states. This disease cohort consists of donors with active inflammatory bowel disease (IBD), a chronic inflammatory condition of the gastrointestinal (GI) tract, where immune cell organisation within the tissue plays a central role in disease pathogenesis.
Inflammatory bowel disease (IBD) is an umbrella term that includes two main GI tract disorders with different characteristics: Crohn’s disease (CD) and ulcerative colitis (UC).
Spatial transcriptomic technologies enable gene expression to be measured at subcellular resolution while preserving the tissue architecture, allowing researchers to uncover spatial patterns and cell-cell interactions that would be lost in conventional bulk or single-cell RNA-seq data.
4.3 The experimental design and cohorts
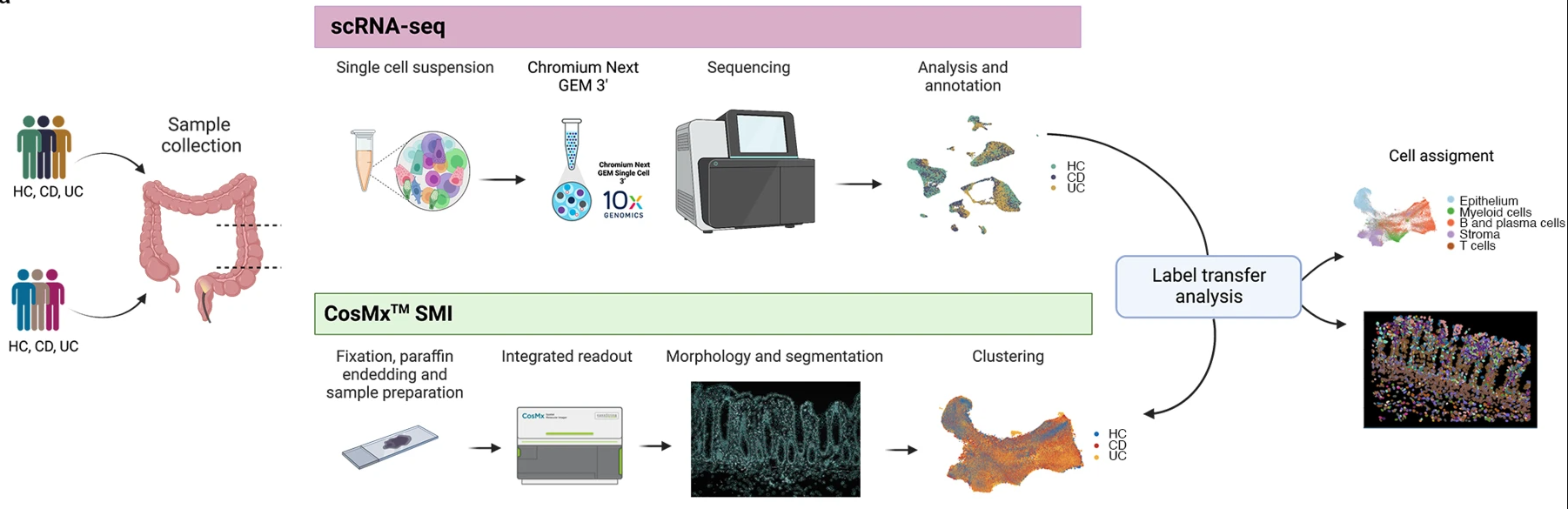
Fig 1a from Garrido-Trigo et al. (2023).
The study included 9 CosMx slides of colonic biopsies, from a total of 9 donors:
| Cohorts | Abbreviated | Number of samples |
|---|---|---|
| Healthy controls | HC | 3 |
| Ulcerative colitis | UC | 3 |
| Crohn’s disease | CD | 3 |
The study includes three biological replicates per condition (HC, UC, CD). This provides enough variation to capture differences between individuals while maintaining manageable data size for hands-on analysis.
In this workshop, we will focus on the healthy control (HC) and Crohn’s disease (CD) samples.
In the original paper, the CosMX SMI cells were annotated via label transfer from the scRNA data. The spatial transcriptomics data will be processed independent of the scRNA data to demonstrate how differences in cellular composition and spatial organisation can be inferred directly from the spatial CosMx data.
TODO: Include image of plots that will be created in downstream steps.