Transfer from WEB to Artemis HPC
Overview
Teaching: 10 min
Exercises: 10 minQuestions
What should you think about when transferring large datasets?
Objectives
Learn to pull data directly from the internet to HPC
Transfer from WEB to Artemis HPC
You need to get a reference file from an online database to use as input for your analysis. A simple way of retrieving files from the web is using the command line utility ‘wget’ (web-get).
In a web browser, go to https://biomirror.mirror.ac.za/ncbigenomes/Canis_familiaris/CHR_05/ . Right click on the file cfa_ref_CanFam3.1_chr5.fa.gz then select “Copy link address” or “Copy link location”.
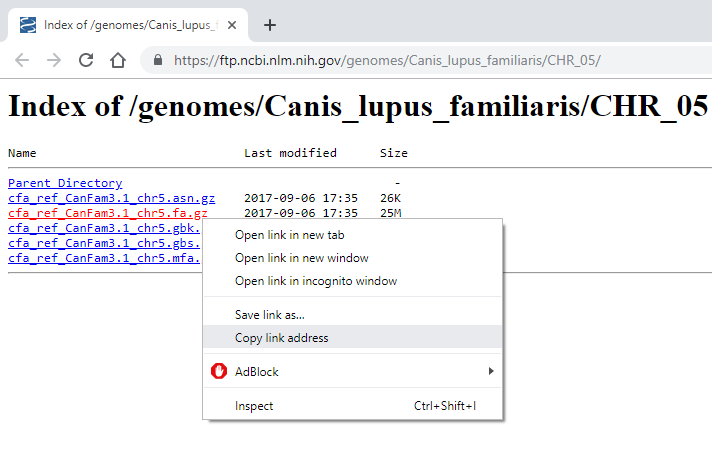
Within your Artemis terminal, enter the wget command, then after a space, paste the URL:
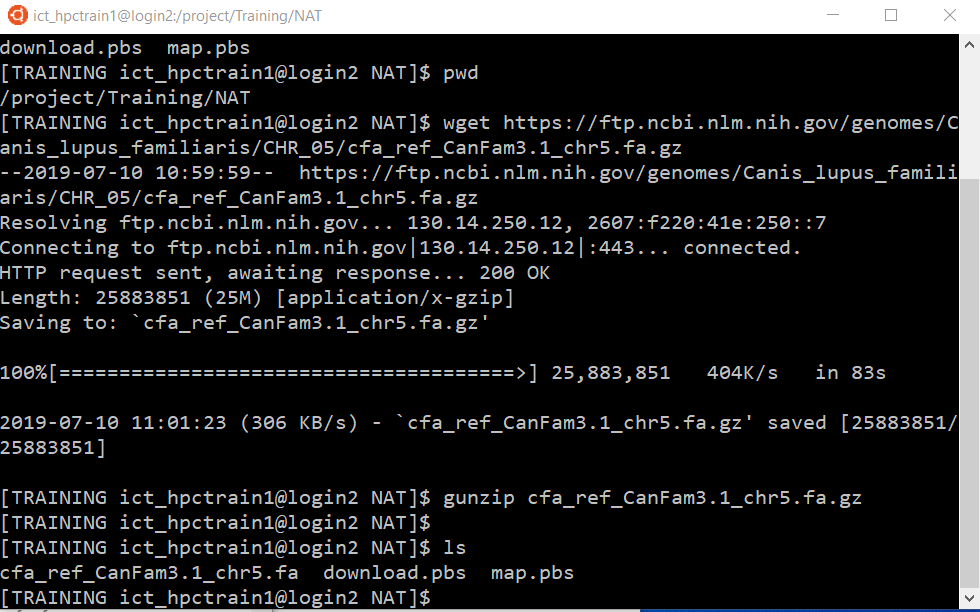
wget https://biomirror.mirror.ac.za/ncbigenomes/Canis_familiaris/CHR_05/cfa_ref_CanFam3.1_chr5.fa.gz
Unzip the file:
gunzip cfa_ref_CanFam3.1_chr5.fa.gz
While wget is very handy for small files, it poses some risks for larger files or large collections of files. Can you think of what these might be?
- Transfers will be aborted if internet or VPN connection is lost, or if your computer sleeps or is shut down, or if there is a network timeout.
Can you think of a solution to this?
- Run the transfer using ‘nohup’ or ‘screen’, or submit the transfer to the cluster using ‘qsub’.
Our next data transfer is a bit larger, so we will use a different method that is more robust.
Transfer from WEB to Artemis HPC: a safer way with ‘dtq’
The raw DNA sequence data for this analysis is hosted online. You could run wget in the foreground as we did for the last data transfer, but we won’t because the files are large.
We will instead ‘wrap’ the wget commands in a PBS script, as you are now familiar with from this morning’s Introduction to the Artemis HPC course. We will submit them to a dedicated data transfer queue on Artemis called dtq which is exempt from fair share. This will be good news to those of you with big datasets to move around 😊
View the script download.pbs with the cat command:
cat download.pbs
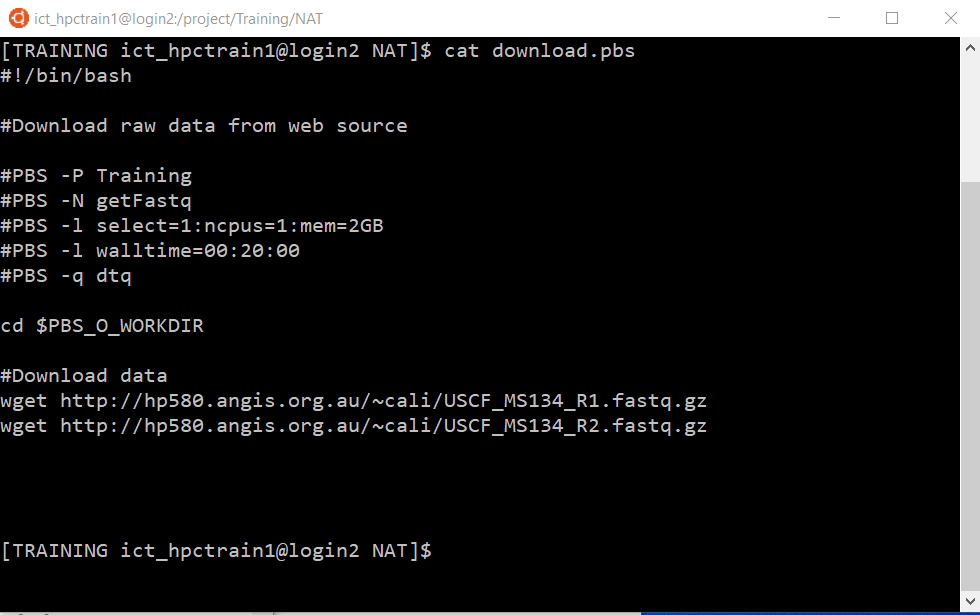
You don’t need to change anything in this script, but note that we have specified to run the job on the dtq queue using the -q flag.
Also note that the data will be downloaded to your current working directory (ie, where you are situated when you run the qsub command). This is because the script changes you to that directory using the PBS environment variable $PBS_O_WORKDIR. If you are not in the correct directory to receive your download, change there now, then submit the transfer with:
qsub download.pbs
Check the status with qstat:
qstat -xu <unikey>
or
qstat -x <jobID>
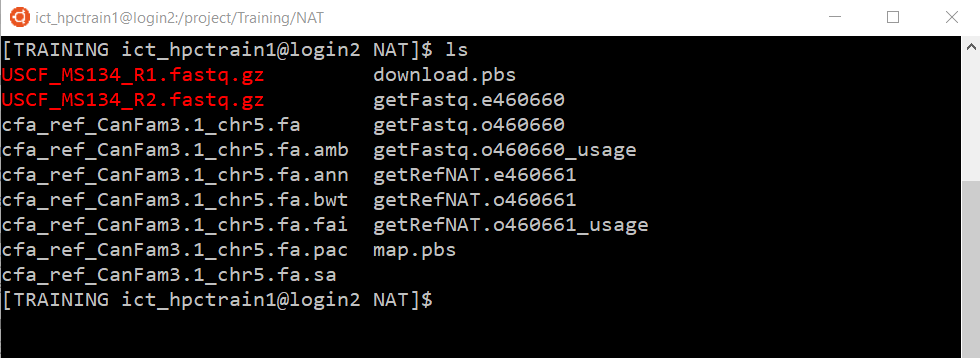
Once your transfer job has completed, confirm that you have the expected files (e.g. ls).
Note that you have the same 3 job log files, like any normal job submitted to Artemis with qsub.
Key Points
Use wget
Recall batch submission on Artemis
Use the dedicated data transfer queue, dtq