Exploring usage of the adjective lemma OBESE
There is interest in exploring statistics around the use of the adjective lemma OBESE around the following research questions:
- Do tabloids use OBESE more than broadsheets?
- Or do broadsheets use OBESE more than tabloids?
- Or is there no discernible pattern of use?
- Is there any newspaper that uses OBESE more than others?
- Any year where OBESE is the most frequent/least frequent?
Additional question:
- Is there a difference of use by primary topic?
Executive summary
1. Tabloids use the adjective lemma OBESE more frequently than broadsheets.
More specifically, a Welch Two Sample t-test testing the difference between the frequency in broadsheets per 1000 words and frequency in tabloids (mean of broadsheets = 3.16, mean of tabloids = 5.59) suggests that the effect is negative, statistically significant, and small (difference = -2.43, 95% CI [-2.63, -2.23], t(10062.88) = -24.01, p < .001; Cohen’s d = -0.46, 95% CI [-0.50, -0.42]).
The total number of uses of OBESE we observe is higher in tabloids and right leaning-publications, and and lower in broadsheets and left-leaning publications than we would expect based on the word count in these subcorpora (p < 0.001).
The number of articles with use of OBESE we observe is not different between tabloids and broadsheets, or between left and right leaning publications.
2. Tabloids have shorter article lengths than broadsheets.
- More specifically, using a Welch Two Sample t-test comparing the mean word count of articles from broadsheets and tabloids (mean of broadsheet = 778.84, mean of tabloid = 485.42) suggests that the effect is positive, statistically significant, and medium (difference = 293.42, 95% CI [273.82, 313.02], t(7348.69) = 29.34, p < .001; Cohen’s d = 0.60, 95% CI [0.56, 0.65])
3. Investigating the data by source and year revealed that these variables explained a small amount of variance in the data, with OBESE being used less frequently in the Age, Australian, Canberra Times and Sydney Morning Herald relative to the Advertiser, while in the Hobart Mercury and Northern Territorian it was used somewhat more frequently than in the Advertiser. Use of the adjective lemma OBESE decreased with time in the corpus.
4. Significant differences in the use of OBESE were observed in articles on different topics, with articles annotated as “Awards” and “Bio-medical Research” using more instances per 1000 words than articles discussing politics, schooling, transport and commuting.
5. Significant differences in article topics were also observed between tabloids and broadsheets, with the topics “ChildrenParents”, “NutritionStudy”, “WomenPregnancy” and “FitnessExercise” approximately 3x more frequently reported on in tabloids than broadsheets, in contrast to topics like “Politics” and “SportsDoping”, which were approximately evenly represented between the two media types.
Data source
CQPweb data was provided. To calculate normalised frequency, we divide the number of observations from CQPweb by the word count in each article as calculated in Python, and multiple by 1000.
Code
adj_obese <- read_cqpweb("aoc_all_obese_tagadjlemma.txt")
metadata <- read_csv(here("100_data_raw", "corpus_cqpweb_metadata.csv"))
additional_source_metadata <- read_csv(here("100_data_raw", "addition_source_metadata.csv"))
topic_labels <- read_csv(here("100_data_raw", "topic_labels.csv"))
metadata_full <- inner_join(inner_join(metadata,
topic_labels,
by = c("dominant_topic" = "topic_number")),
additional_source_metadata)
obese_annotated <- inner_join(
adj_obese, metadata_full, by = c("text" = "article_id")) %>%
mutate(frequency = 10^3*no_hits_in_text/wordcount_total)|>
rename(article_id = text)We group articles into tabloids and broadsheets, and by orientation, in the following manner:
| source | source_type | orientation |
|---|---|---|
| Advertiser | tabloid | right |
| Australian | broadsheet | right |
| NorthernT | tabloid | right |
| CourierMail | tabloid | right |
| Age | broadsheet | left |
| SydHerald | broadsheet | left |
| Telegraph | tabloid | right |
| WestAus | tabloid | right |
| CanTimes | broadsheet | left |
| HeraldSun | tabloid | right |
| HobMercury | tabloid | right |
| BrisTimes | broadsheet | left |
Tabloid vs broadsheet
What is the distribution of OBESE in articles?
Code
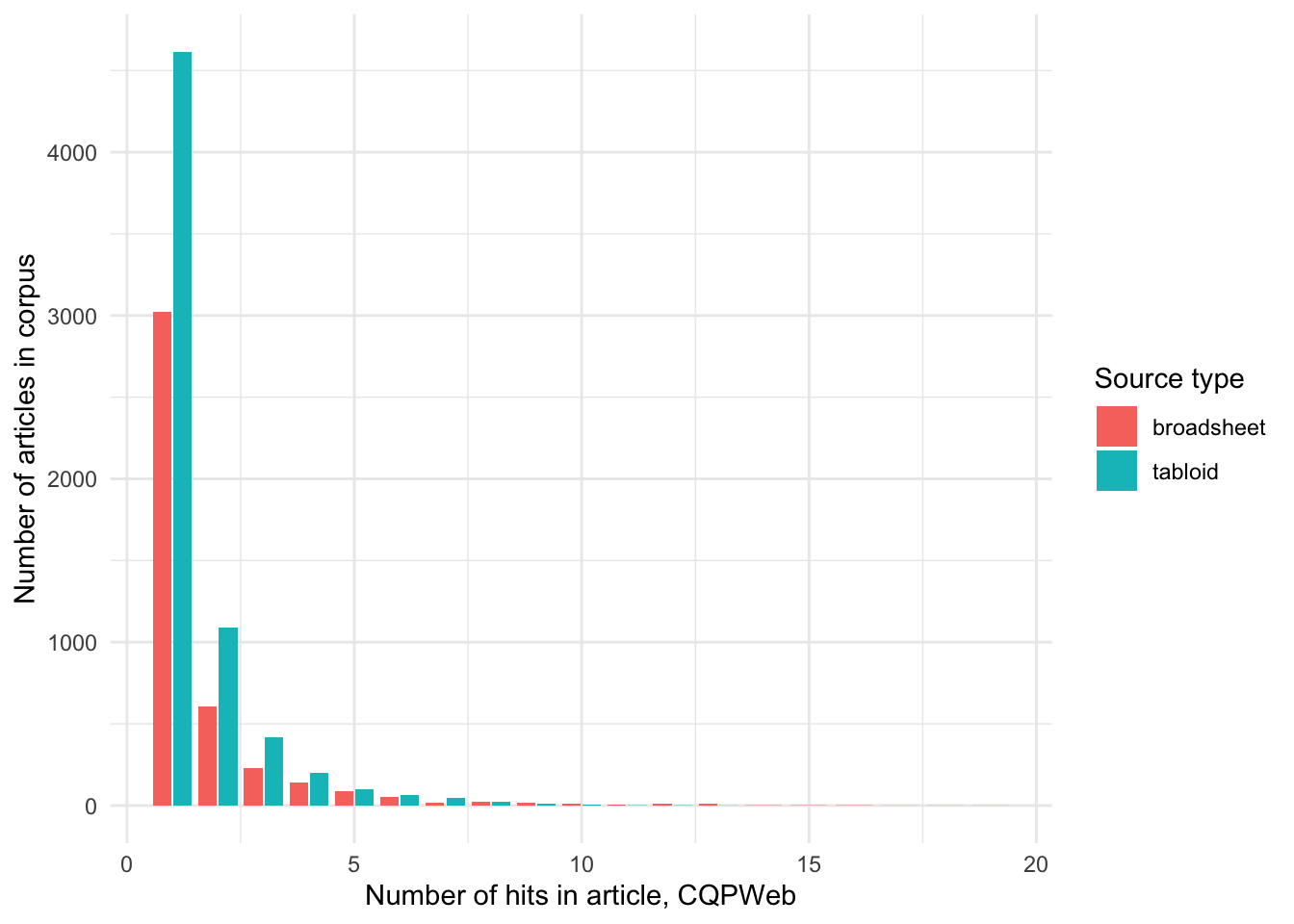
How is this usage distributed by year (number of articles in corpus)?
Code
| source | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Advertiser | 130 | 144 | 115 | 127 | 125 | 158 | 122 | 127 | 100 | 96 | 77 | 75 | 1396 |
| Age | 140 | 85 | 107 | 85 | 102 | 94 | 88 | 135 | 93 | 90 | 63 | 44 | 1126 |
| Australian | 115 | 84 | 68 | 76 | 58 | 47 | 61 | 33 | 35 | 34 | 62 | 36 | 709 |
| CanTimes | 71 | 85 | 70 | 79 | 72 | 88 | 98 | 88 | 54 | 77 | 65 | 26 | 873 |
| CourierMail | 167 | 144 | 125 | 110 | 97 | 118 | 122 | 108 | 100 | 85 | 118 | 74 | 1368 |
| HeraldSun | 196 | 201 | 171 | 183 | 138 | 157 | 121 | 95 | 84 | 89 | 95 | 75 | 1605 |
| HobMercury | 75 | 59 | 56 | 57 | 60 | 63 | 27 | 33 | 32 | 32 | 43 | 38 | 575 |
| NorthernT | 49 | 54 | 45 | 30 | 33 | 31 | 27 | 30 | 18 | 20 | 22 | 12 | 371 |
| SydHerald | 158 | 126 | 129 | 130 | 130 | 124 | 101 | 142 | 105 | 110 | 98 | 81 | 1434 |
| WestAus | 114 | 96 | 90 | 98 | 95 | 77 | 83 | 44 | 53 | 27 | 16 | 6 | 799 |
| BrisTimes | 0 | 0 | 0 | 0 | 0 | 1 | 9 | 37 | 5 | 7 | 19 | 22 | 100 |
| Telegraph | 0 | 0 | 0 | 0 | 0 | 0 | 93 | 52 | 86 | 95 | 79 | 60 | 465 |
| Total | 1215 | 1078 | 976 | 975 | 910 | 958 | 952 | 924 | 765 | 762 | 757 | 549 | 10821 |
We can see that apart from the Brisbane times and Daily Telegraph, there are articles using OBESE every year and in every publication. We will filter out these two sources.
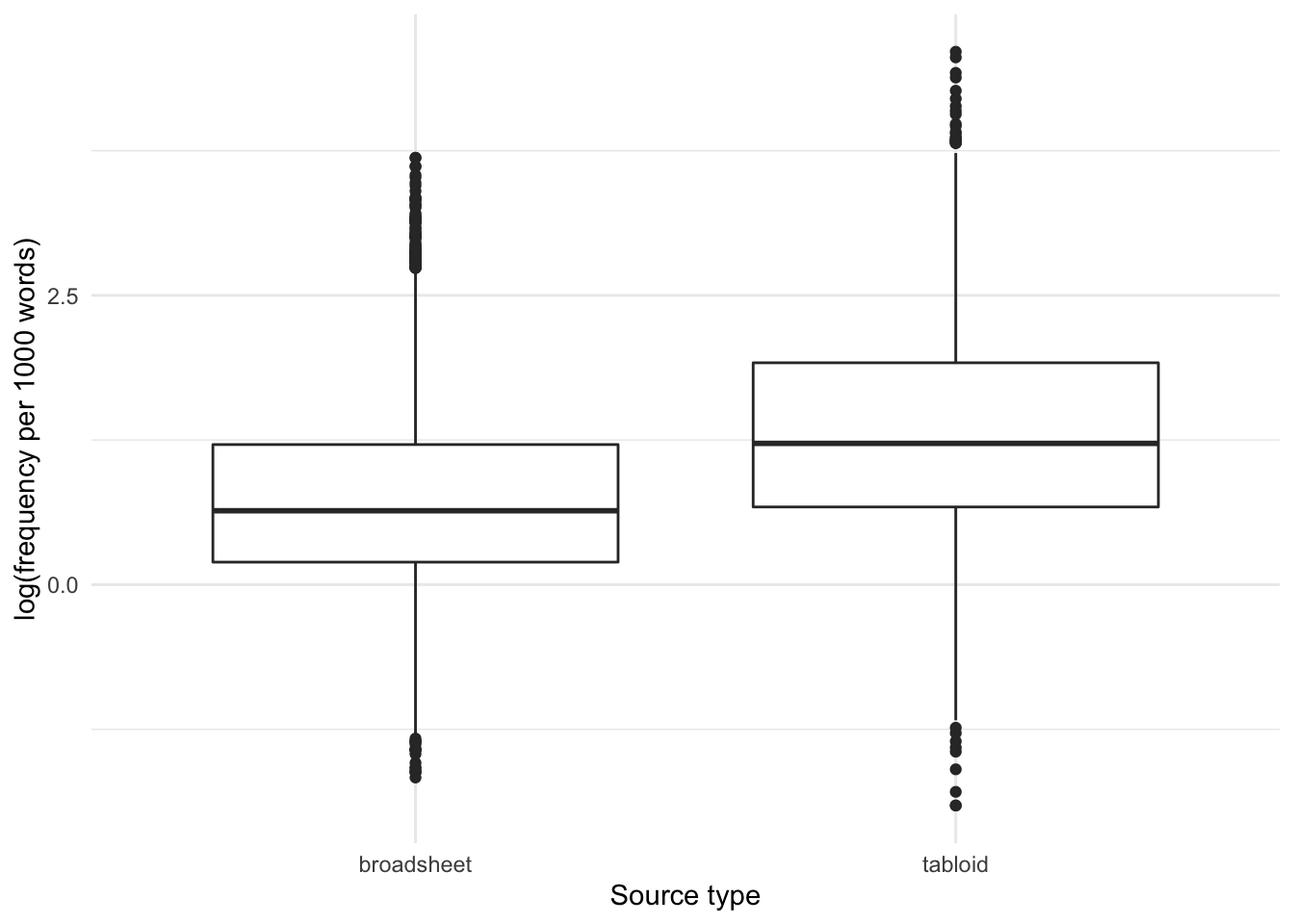
How is the frequency (per thousand words) of the usage of OBESE distributed by tabloids/broadsheets?
Code
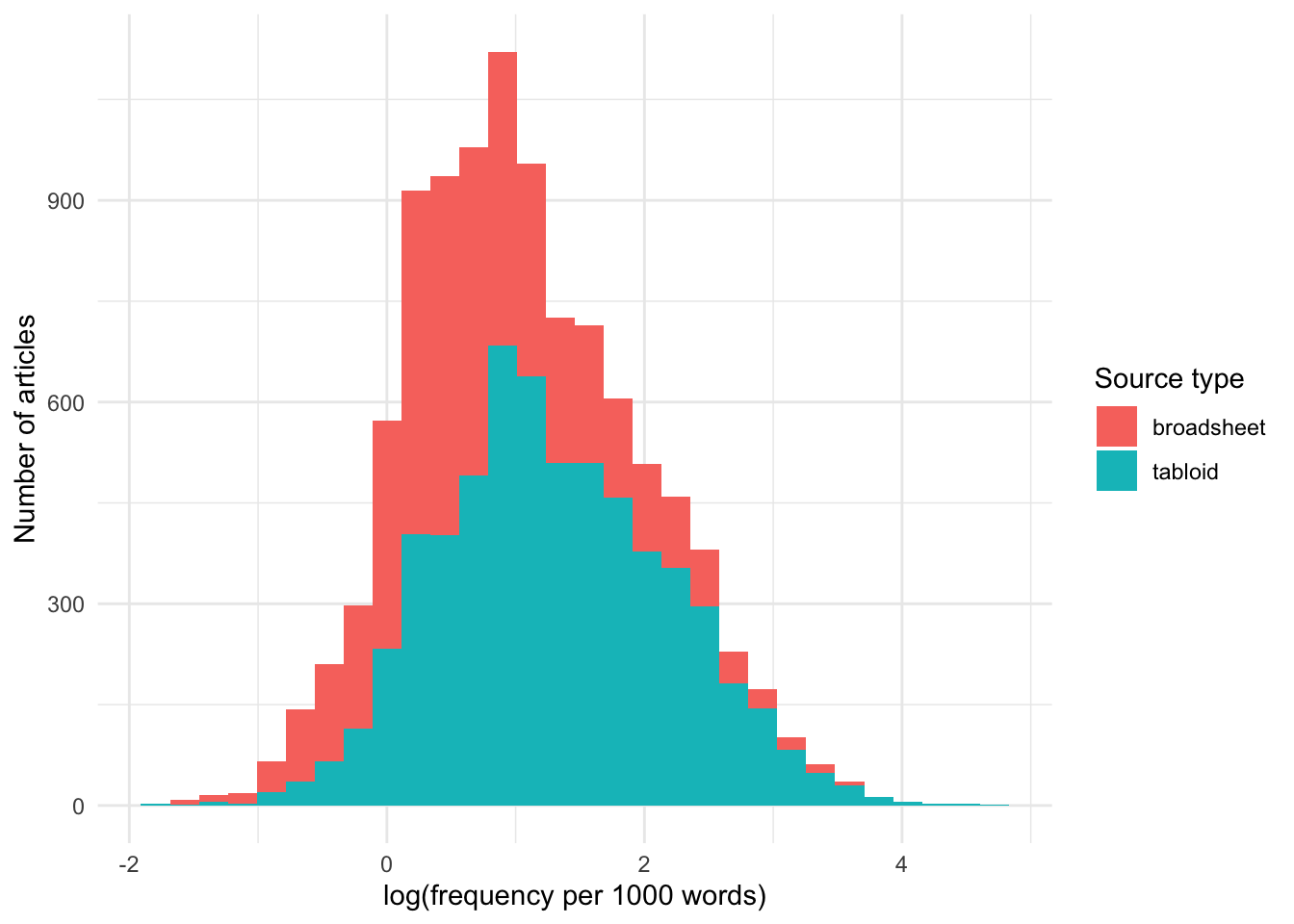
And let’s also use a histogram to look at the distribution:
Code
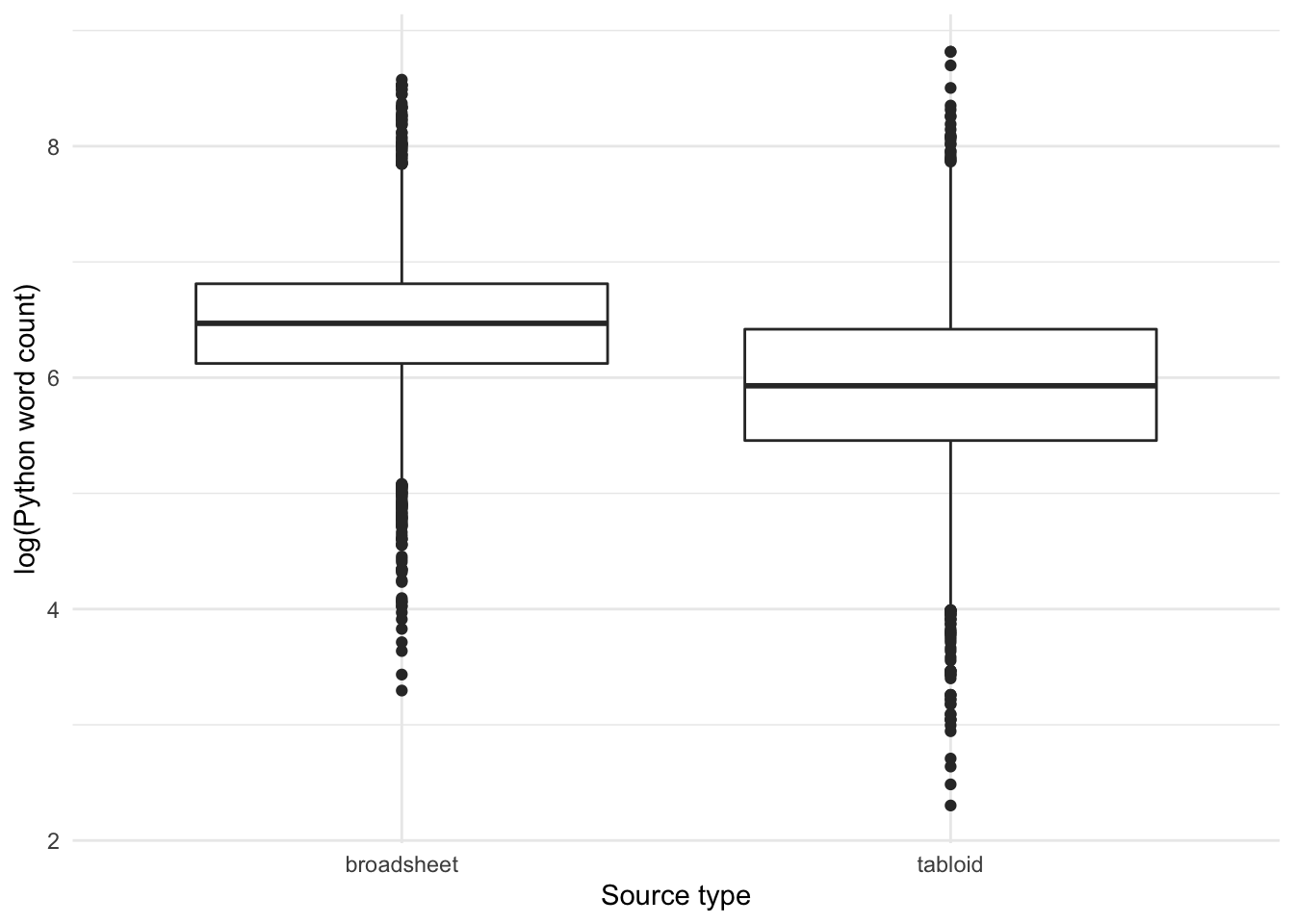
Note that broadsheets have somewhat longer texts than tabloids:
Code
Let’s use a histogram to look at the distribution in more detail:
Code
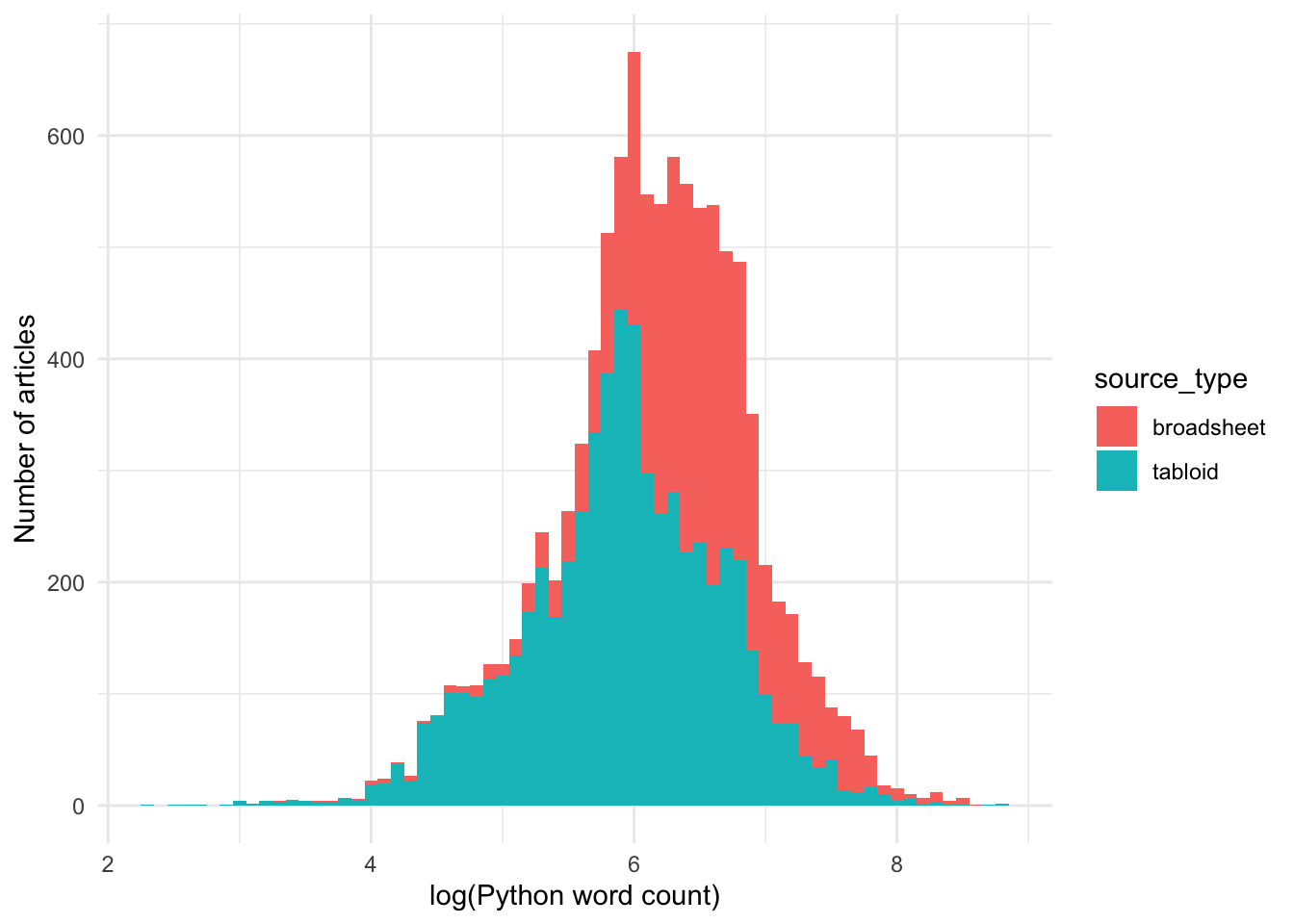
The log-transformed word count data is approximately normally distributed.
Let’s see if the difference in length of articles using the adjective lemma OBESE in tabloids and broadsheets is significant?
Code
wordcount_broadsheet <- obese_annotated_filtered[obese_annotated_filtered$source_type == "broadsheet","wordcount_total", drop = TRUE]
wordcount_tabloid <- obese_annotated_filtered[obese_annotated_filtered$source_type == "tabloid","wordcount_total", drop = TRUE]
report::report(t.test(wordcount_broadsheet,wordcount_tabloid))The Welch Two Sample t-test testing the difference between wordcount_broadsheet and wordcount_tabloid (mean of x = 778.84, mean of y = 485.42) suggests that the effect is positive, statistically significant, and medium (difference = 293.42, 95% CI [273.82, 313.02], t(7348.69) = 29.34, p < .001; Cohen’s d = 0.60, 95% CI [0.56, 0.65])
Yes, overall articles that use the adjective lemma OBESE in tabloids are significantly shorter than in broadsheets.
This is supported by a non-parametric test as well:
Code
fp_test(wc1 = wordcount_broadsheet,
wc2 = wordcount_tabloid,
label1 = "broadsheet",
label2 = "tabloid",
dist = mydistribution)
Approximative Two-Sample Fisher-Pitman Permutation Test
data: wc by label (broadsheet, tabloid)
Z = 29.474, p-value < 1e-04
alternative hypothesis: true mu is not equal to 0Let’s see whether this holds overall, across all articles in tabloids vs broadsheets, not just the ones that use the adjective lemma OBESE:
Code
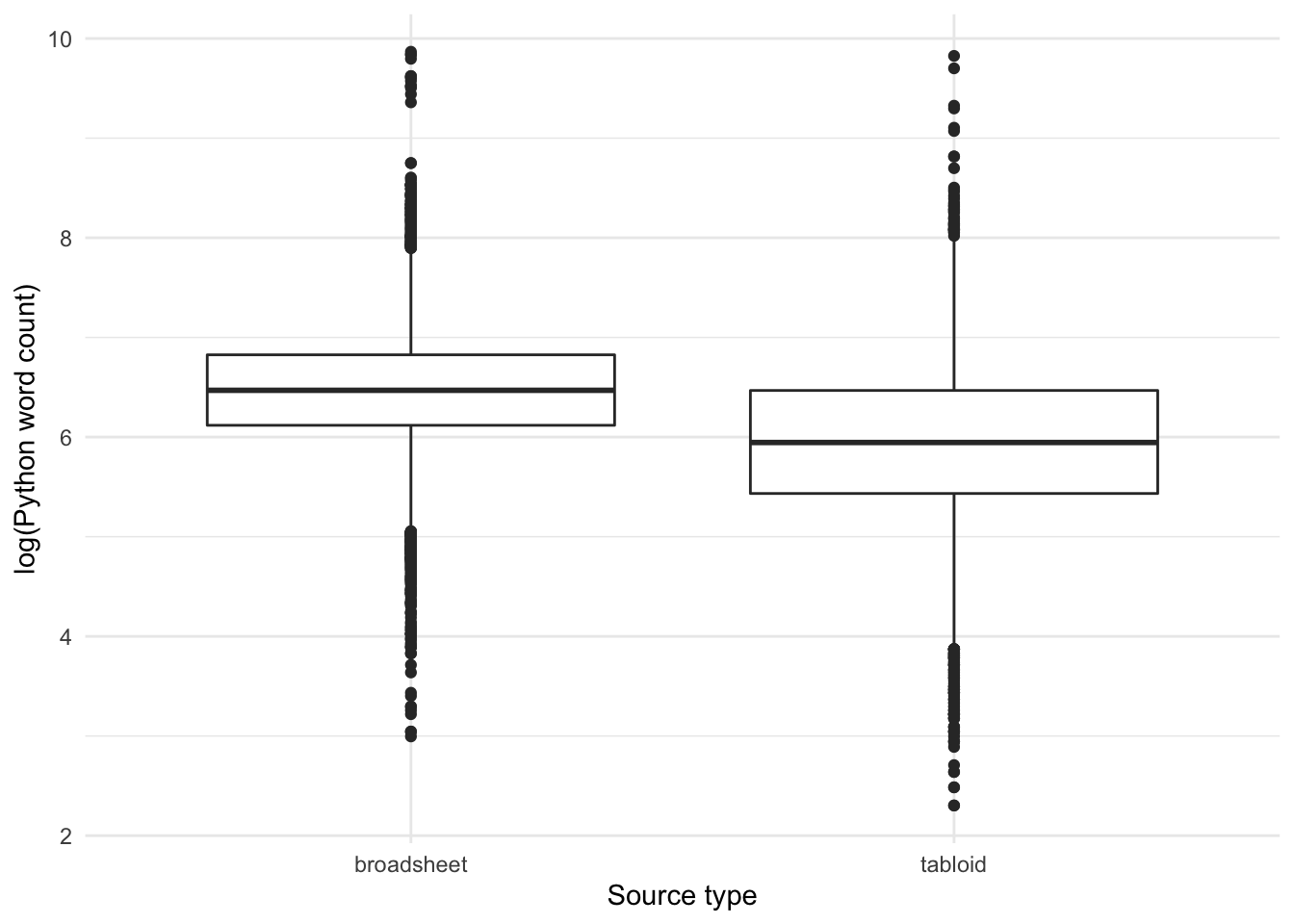
Let’s use a histogram to look at the distribution in more detail:
Code
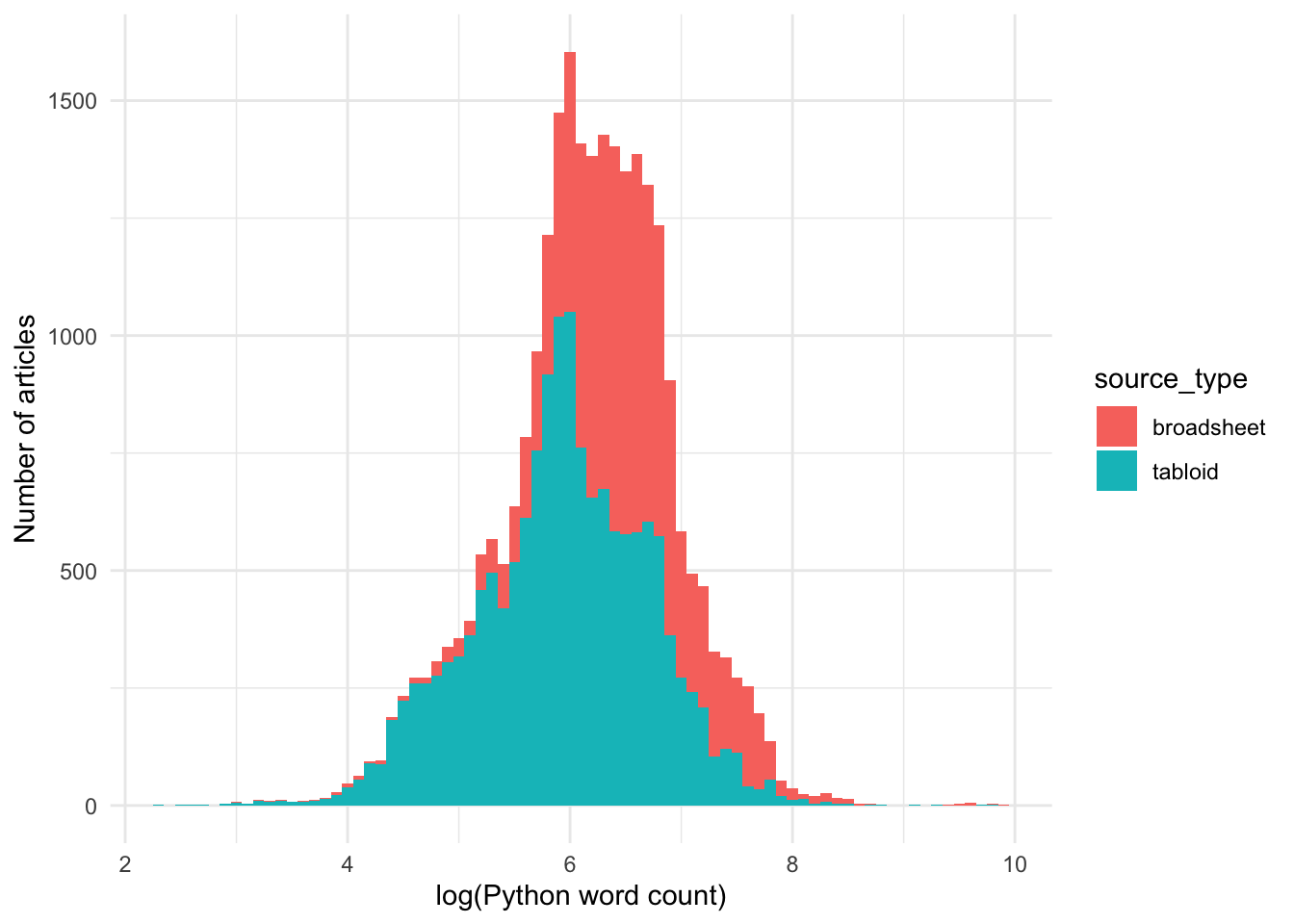
The log-transformed word count data is approximately normally distributed.
Let’s see if the difference in length of articles using the adjective lemma OBESE in tabloids and broadsheets is significant?
Code
The Welch Two Sample t-test testing the difference between wordcount_broadsheet and wordcount_tabloid (mean of x = 810.15, mean of y = 502.44) suggests that the effect is positive, statistically significant, and small (difference = 307.71, 95% CI [291.01, 324.42], t(16482.54) = 36.11, p < .001; Cohen’s d = 0.47, 95% CI [0.45, 0.50])
Yes, we can see that across the entire dataset, the word count of articles published in broadsheets are longer than that in tabloids.
This is supported by a non-parametric test as well:
Code
fp_test(wc1 = wordcount_broadsheet,
wc2 = wordcount_tabloid,
label1 = "broadsheet",
label2 = "tabloid",
dist = mydistribution)
Approximative Two-Sample Fisher-Pitman Permutation Test
data: wc by label (broadsheet, tabloid)
Z = 37.999, p-value < 1e-04
alternative hypothesis: true mu is not equal to 0If we then test the difference between the frequency of the adjective lemma OBESE in tabloids and broadsheets, we can see that a higher frequency per 1000 words is detected for usage of the adjective lemma OBESE in tabloids than broadsheets.
Code
frequency_broadsheet <- obese_annotated_filtered[obese_annotated_filtered$source_type == "broadsheet","frequency", drop = T]
frequency_tabloid <- obese_annotated_filtered[obese_annotated_filtered$source_type == "tabloid","frequency", drop = T]
report::report(t.test(frequency_broadsheet, frequency_tabloid))The Welch Two Sample t-test testing the difference between frequency_broadsheet and frequency_tabloid (mean of x = 3.16, mean of y = 5.59) suggests that the effect is negative, statistically significant, and small (difference = -2.43, 95% CI [-2.63, -2.23], t(10062.88) = -24.01, p < .001; Cohen’s d = -0.46, 95% CI [-0.50, -0.42])
So, yes, the frequency of use of the adjective lemma OBESE is higher in tabloids than in broadsheets.
This is supported by a non-parametric test as well:
Code
fp_test(wc1 = frequency_broadsheet,
wc2 = frequency_tabloid,
label1 = "broadsheet",
label2 = "tabloid",
dist = mydistribution)
Approximative Two-Sample Fisher-Pitman Permutation Test
data: wc by label (broadsheet, tabloid)
Z = -21.36, p-value < 1e-04
alternative hypothesis: true mu is not equal to 0Let’s use bootstrapping to see if the raw frequencies of usage of OBESE are different?
Code
broadsheet_counts <- NULL
tabloid_counts <- NULL
for (i in 1:10000) {
x <- mean(sample({obese_annotated_filtered %>%
filter(source_type == "broadsheet") %>%
pull(no_hits_in_text)}, 1000, replace = FALSE))
y <- mean(sample({obese_annotated_filtered %>%
filter(source_type == "tabloid") %>%
pull(no_hits_in_text)}, 1000, replace = FALSE))
broadsheet_counts <- c(broadsheet_counts, x)
tabloid_counts <- c(tabloid_counts, y)
}
counts_comparison <- data.frame(
mean_sample = c(broadsheet_counts, tabloid_counts),
source_type = c(
rep("broadsheet", length(broadsheet_counts)),
rep("tabloid", length(tabloid_counts))))
counts_comparison %>%
ggplot(aes(x = mean_sample, fill = source_type)) +
geom_histogram()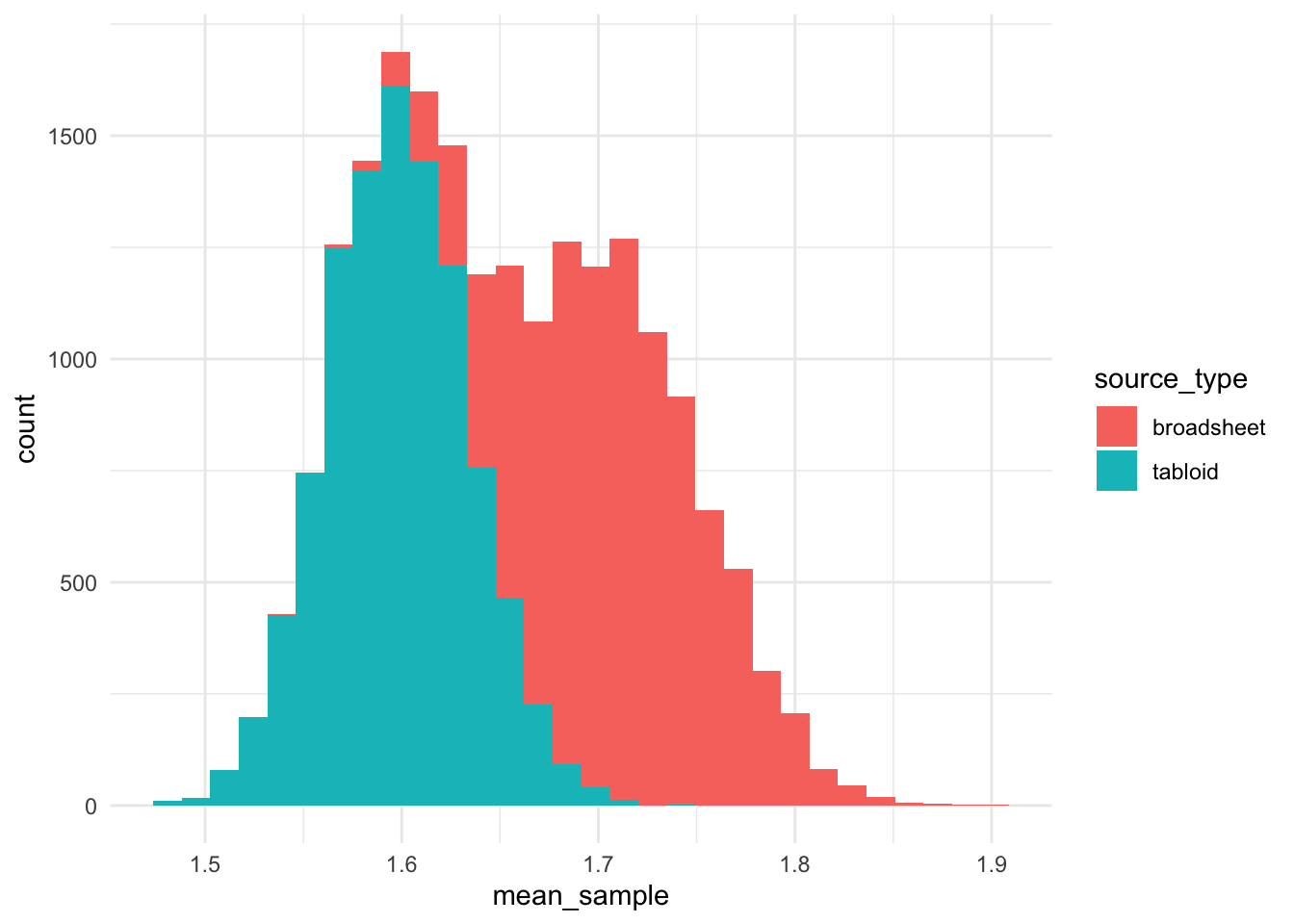
It is interesting that the distribution of usage of OBESE is ~1.6 uses per article in tabloid publications, while the distribution for broadsheets was bimodal.
The Welch Two Sample t-test testing the difference between broadsheet_counts and tabloid_counts (mean of x = 1.71, mean of y = 1.60) suggests that the effect is positive, statistically significant, and large (difference = 0.11, 95% CI [0.11, 0.11], t(18734.64) = 186.07, p < .001; Cohen’s d = 2.63, 95% CI [2.89, 2.89])
This is supported by a non-parametric test as well:
Code
fp_test(wc1 = broadsheet_counts,
wc2 = tabloid_counts,
label1 = "broadsheet",
label2 = "tabloid",
dist = mydistribution)
Approximative Two-Sample Fisher-Pitman Permutation Test
data: wc by label (broadsheet, tabloid)
Z = 112.59, p-value < 1e-04
alternative hypothesis: true mu is not equal to 0So the COUNT of instances of the adjective lemma OBESE is HIGHER in broadsheets than tabloids - the opposite result of when we consider the frequency. This is likely to be due to broadsheets having much longer articles than tabloids.
Let’s look at the trend over time:
Code
obese_annotated_filtered %>%
ggplot(aes(x = as.factor(year),
y = log(frequency),
fill = year)) +
geom_jitter(alpha = 0.2) +
geom_smooth(aes(group = source_type), col = "blue", method = "loess") +
geom_hline(yintercept = 1, col = "red", lty = 3, size = 3) +
facet_wrap(~source_type) +
theme(axis.text.x =
element_text(angle = 90, vjust = 0.5, hjust=1),
legend.position = "NA") +
labs(
x = "Year",
y = "log(frequency per 1000 words)"
)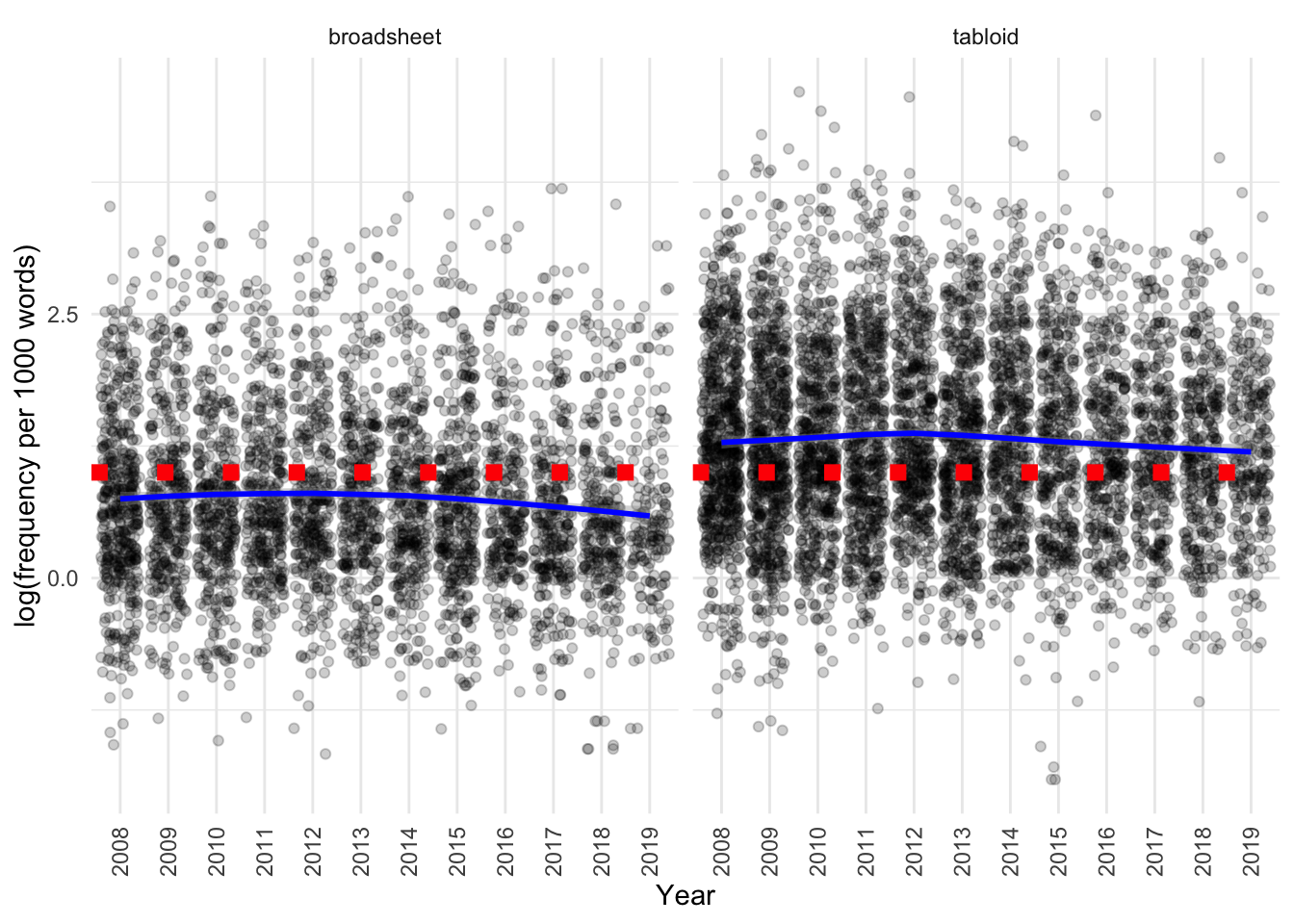
We can see that there is a difference:
- between tabloids and broadsheets, as characterised above
- usage subtly decreases in both over time
Comparing observed to normalised to subcorpus size data
We can investigate the prevalence of the use of OBESE using goodness of fit tests, comparing the distribution in:
- tabloids vs broadsheets
- left and right leaning publications
We can do this by looking at:
- the total number of instances in each subcorpus, normalised to the total number of words in each subcorpus
- the number of articles that feature this language type, normalised to the total number of articles in each subcorpus
Tabloids vs broadsheets
The total number of uses of OBESE we observe is higher in tabloids and lower in broadsheets than we would expect based on the word count in these subcorpora (p < 0.001).
Code
chisq_instances_wc_normalised(obese_annotated_filtered, metadata_full, source_type) |> kable()| variable | value |
|---|---|
| method | Chi-squared test for given probabilities |
| parameter | 1 |
| statistic | 778.4111 |
| p.value | 2.666527e-171 |
| broadsheet_observed | 7071 |
| broadsheet_expected | 8878.82882136884 |
| tabloid_observed | 9773 |
| tabloid_expected | 7965.17117863116 |
The number of articles with use of OBESE we observe is not different between tabloids and broadsheets.
Code
chisq_articles_totalart_normalised(obese_annotated_filtered, metadata_full, source_type) |> kable()| variable | value |
|---|---|
| method | Chi-squared test for given probabilities |
| parameter | 1 |
| statistic | 1.012291 |
| p.value | 0.3143545 |
| broadsheet_observed | 4142 |
| broadsheet_expected | 4192.09050949815 |
| tabloid_observed | 6114 |
| tabloid_expected | 6063.90949050185 |
Left vs right-leaning publications
The total number of uses of OBESE we observe is higher in right and lower in left-leaning publications than we would expect based on the word count in these subcorpora (p < 0.001).
Code
chisq_instances_wc_normalised(obese_annotated_filtered, metadata_full, orientation) |> kable()| variable | value |
|---|---|
| method | Chi-squared test for given probabilities |
| parameter | 1 |
| statistic | 336.0413 |
| p.value | 4.645282e-75 |
| left_observed | 5938 |
| left_expected | 7113.11347665779 |
| right_observed | 10906 |
| right_expected | 9730.88652334221 |
The number of articles that use OBESE we observe is not different between left and right leaning publications.
Code
chisq_articles_totalart_normalised(obese_annotated_filtered, metadata_full, orientation) |> kable()| variable | value |
|---|---|
| method | Chi-squared test for given probabilities |
| parameter | 1 |
| statistic | 0.03741104 |
| p.value | 0.8466306 |
| left_observed | 3433 |
| left_expected | 3423.7627183427 |
| right_observed | 6823 |
| right_expected | 6832.2372816573 |
Differences in usage by source
Is there a difference in the usage of OBESE by source?
Code
obese_annotated_filtered %>%
ggplot(aes(x = reorder(source,frequency),
y = log(frequency),
fill = source_type)) +
geom_boxplot() +
theme(axis.text.x =
element_text(angle = 90, vjust = 0.5, hjust=1),
legend.position = "NA")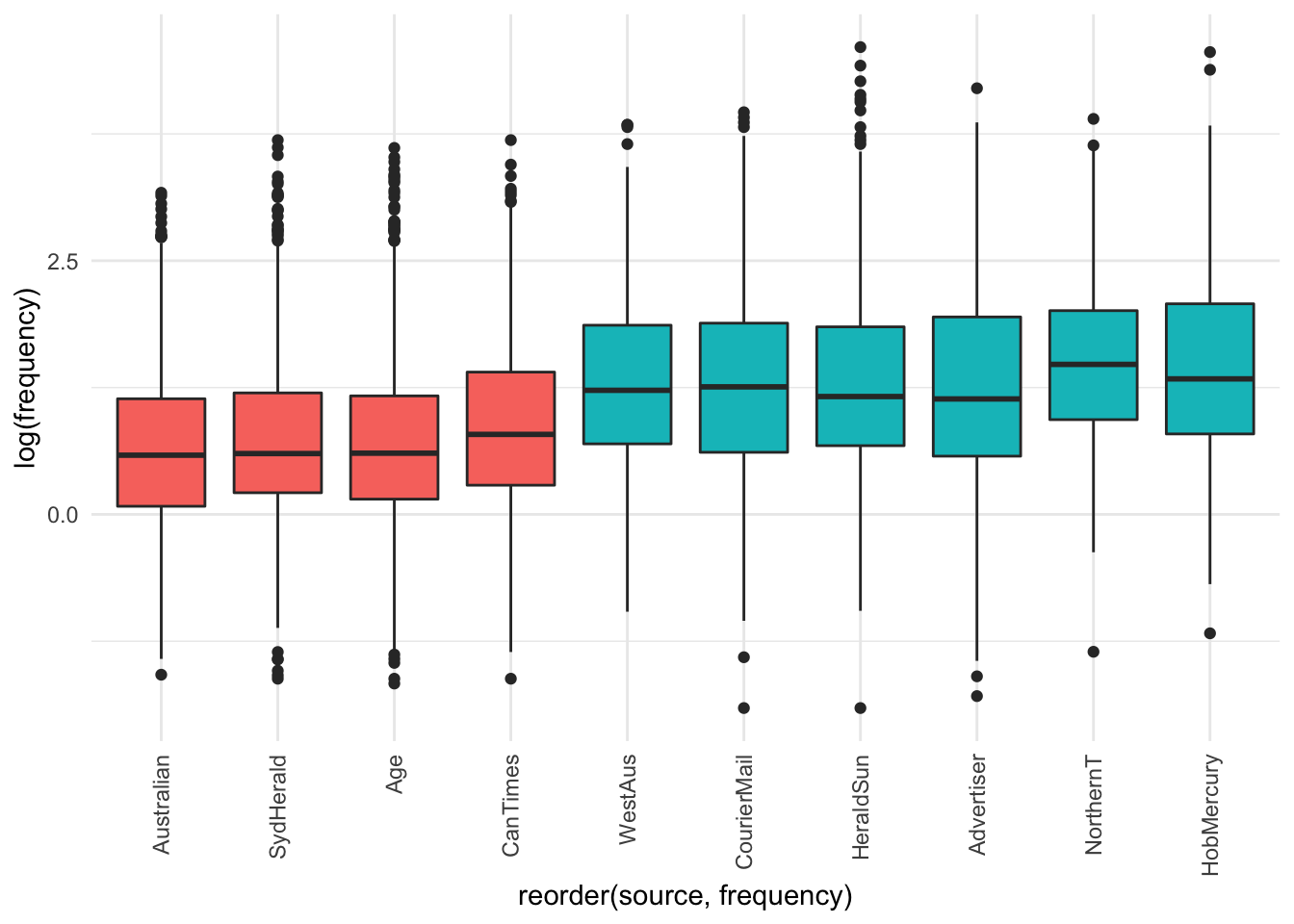
The visualisation suggests there is not - only the differences observed above for tabloids vs broadsheets.
What are the means and standard deviations of the frequency by source?
Code
| source | mean | median | sd | type |
|---|---|---|---|---|
| Australian | 2.817048 | 1.792115 | 3.175300 | broadsheet |
| SydHerald | 3.072108 | 1.821494 | 3.657049 | broadsheet |
| Age | 3.154595 | 1.828154 | 4.060922 | broadsheet |
| CanTimes | 3.569830 | 2.200220 | 4.025436 | broadsheet |
| WestAus | 5.292246 | 3.397508 | 5.496833 | tabloid |
| CourierMail | 5.351845 | 3.514949 | 5.800857 | tabloid |
| HeraldSun | 5.493390 | 3.194888 | 6.965477 | tabloid |
| Advertiser | 5.565510 | 3.120132 | 6.236993 | tabloid |
| NorthernT | 6.251933 | 4.385965 | 6.097086 | tabloid |
| HobMercury | 6.464202 | 3.802281 | 8.143909 | tabloid |
It seems that within the different sources among broadsheets and tabloids there is not much difference among the frequency of use of the adjective lemma OBESE.
Differences in usage by year
Is there any year when OBESE is used more frequently than others?
Code
obese_annotated_filtered %>%
ggplot(aes(x = reorder(year,frequency),
y = log(frequency),
fill = year)) +
geom_boxplot() +
theme(axis.text.x =
element_text(angle = 90, vjust = 0.5, hjust=1),
legend.position = "NA")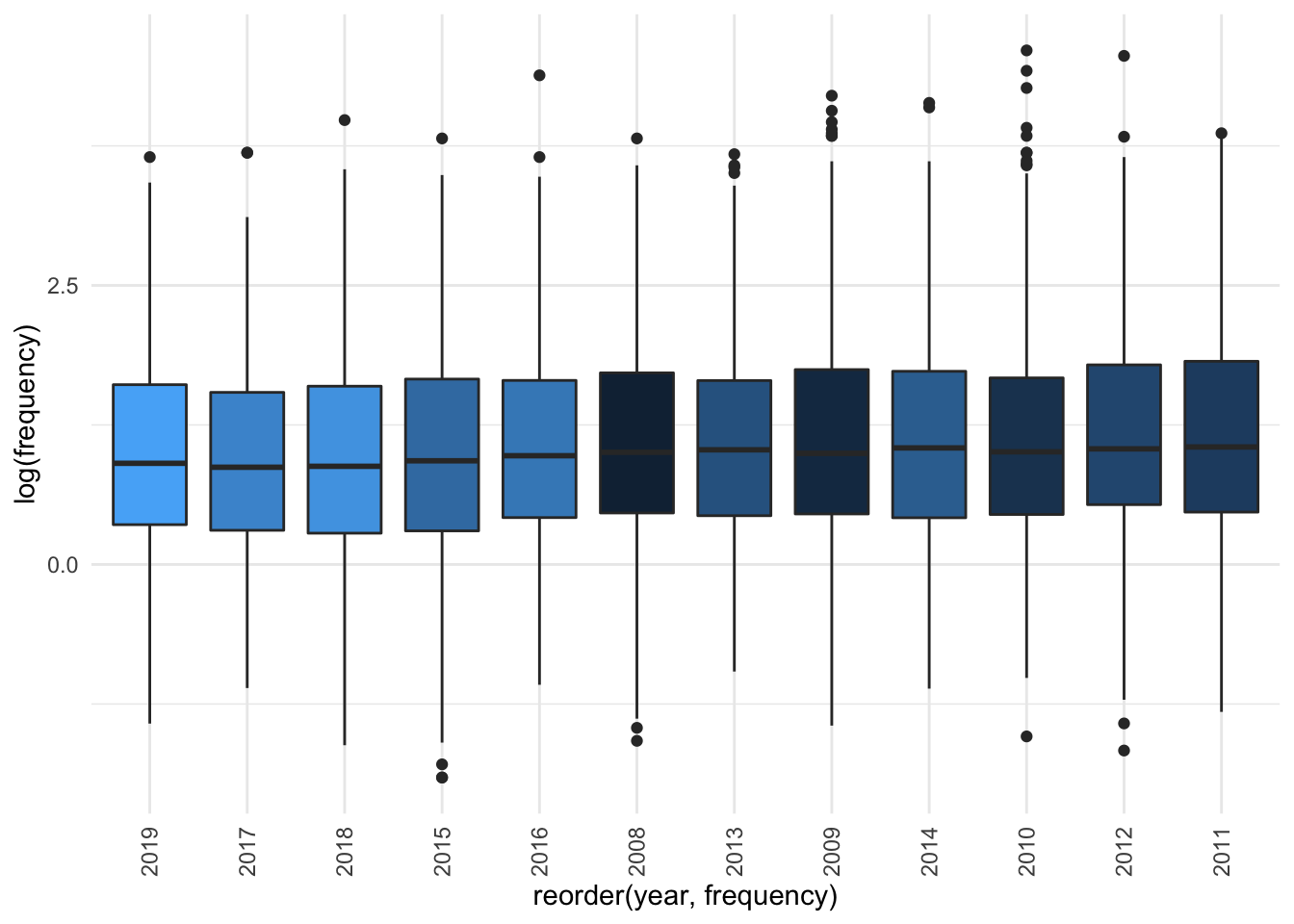
Based on the visualisation it seems not. If we separate out by source there also doesn’t seem to be much difference. If we use a jitter plot to visualise the data, then fit a smoothing line and compare with the line of “no change” (dashed red line), we can see that there really isn’t a very strong difference by source and year:
Code
obese_annotated_filtered %>%
ggplot(aes(x = as.factor(year),
y = log(frequency),
fill = year)) +
geom_jitter(alpha = 0.2) +
geom_smooth(aes(group = source), col = "blue", method = "loess") +
geom_hline(yintercept = 1, col = "red", lty = 3, size = 3) +
facet_wrap(~source) +
theme(axis.text.x =
element_text(angle = 90, vjust = 0.5, hjust=1),
legend.position = "NA") +
labs(
x = "Year",
y = "log(frequency per 1000 words)"
)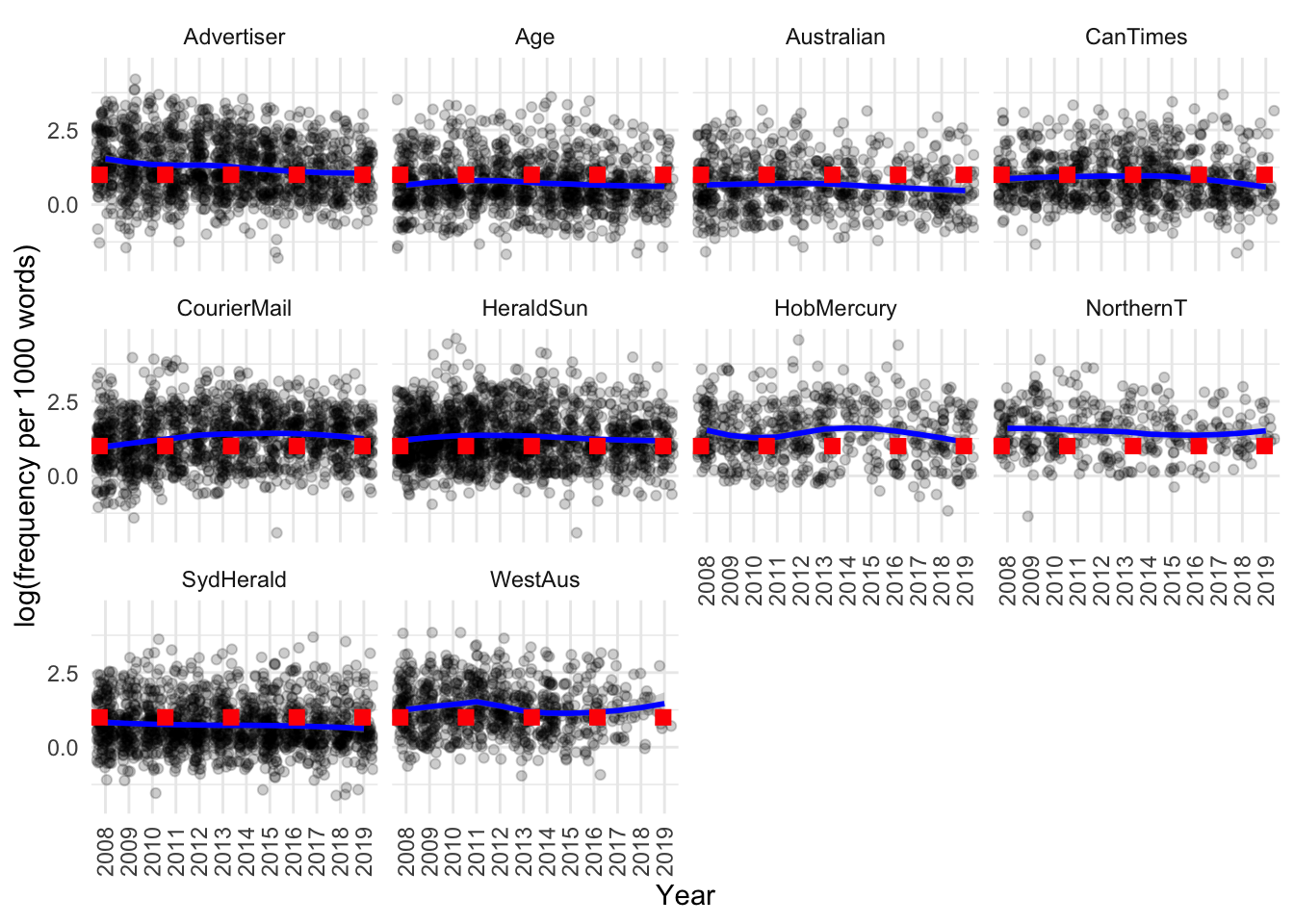
The distributions each year also look quite similar:
Code
obese_annotated_filtered %>%
ggplot(aes(x = reorder(year,frequency),
y = log(frequency),
fill = year)) +
geom_violin() +
theme(axis.text.x =
element_text(angle = 90, vjust = 0.5, hjust=1),
legend.position = "NA") +
labs(x = "Year")
Code
| year | mean | median | sd |
|---|---|---|---|
| 2019 | 3.817842 | 2.475248 | 3.971667 |
| 2017 | 3.866365 | 2.389490 | 4.300584 |
| 2018 | 3.966531 | 2.409639 | 4.616181 |
| 2015 | 4.335848 | 2.531646 | 4.950555 |
| 2016 | 4.471581 | 2.649011 | 5.673864 |
| 2008 | 4.512463 | 2.732240 | 4.882211 |
| 2013 | 4.581882 | 2.793296 | 5.057143 |
| 2009 | 4.801149 | 2.706365 | 6.176861 |
| 2014 | 4.926691 | 2.840909 | 6.018102 |
| 2010 | 4.938758 | 2.743489 | 7.376403 |
| 2012 | 5.065254 | 2.816924 | 6.234889 |
| 2011 | 5.128600 | 2.865330 | 6.218089 |
Differences in usage by source type, source and year
We can also try to simultaneously model differences by source type, source and year.
As expected, a model that includes the source type (tabloid vs broadsheet) gives a better fit than one that does not:
Code
obese_annotated_filtered$scaled_year <- scale(
obese_annotated_filtered$year, scale = F)
library(lme4)
# base model
m_0_base <- lm(log(frequency) ~ 1,
data = obese_annotated_filtered)
# with source type
m_0_sourcetype <- lm(log(frequency) ~ source_type,
data = obese_annotated_filtered)
# with year
m_0_year <- lm(log(frequency) ~ scaled_year,
data = obese_annotated_filtered)
# with source
m_0_source <- lm(log(frequency) ~ source,
data = obese_annotated_filtered)
# with source and year
# with source
m_0_sourceyear <- lm(log(frequency) ~ source + scaled_year,
data = obese_annotated_filtered)
# compare
rbind({broom::glance(m_0_base) %>%
dplyr::select(-df.residual,- deviance, -nobs) %>%
mutate(model = "1")},
{broom::glance(m_0_sourcetype)%>%
dplyr::select(-df.residual,- deviance, -nobs) %>%
mutate(model = "source_type")},
{broom::glance(m_0_year) %>%
dplyr::select(-df.residual,- deviance, -nobs) %>%
mutate(model = "scaled_year")},
{broom::glance(m_0_source) %>%
dplyr::select(-df.residual,- deviance, -nobs) %>%
mutate(model = "source")},
{broom::glance(m_0_sourceyear) %>%
dplyr::select(-df.residual,- deviance, -nobs) %>%
mutate(model = "sourceyear")}
) %>%
dplyr::select(model, everything()) %>%
arrange(AIC) %>%
kable()| model | r.squared | adj.r.squared | sigma | statistic | p.value | df | logLik | AIC | BIC |
|---|---|---|---|---|---|---|---|---|---|
| sourceyear | 0.0965926 | 0.0957108 | 0.8690119 | 109.53982 | 0e+00 | 10 | -13107.20 | 26238.41 | 26325.24 |
| source | 0.0952110 | 0.0944163 | 0.8696337 | 119.79863 | 0e+00 | 9 | -13115.04 | 26252.08 | 26331.67 |
| source_type | 0.0891430 | 0.0890541 | 0.8722045 | 1003.52945 | 0e+00 | 1 | -13149.32 | 26304.63 | 26326.34 |
| scaled_year | 0.0026045 | 0.0025072 | 0.9126977 | 26.77577 | 2e-07 | 1 | -13614.74 | 27235.48 | 27257.19 |
| 1 | 0.0000000 | 0.0000000 | 0.9138440 | NA | NA | NA | -13628.12 | 27260.23 | 27274.70 |
We can see that the model incorporating source and year provides the best fit for the data, explaining somewhat more variability than that which includes only source type.
Code
anova(m_0_sourcetype, m_0_sourceyear)Analysis of Variance Table
Model 1: log(frequency) ~ source_type
Model 2: log(frequency) ~ source + scaled_year
Res.Df RSS Df Sum of Sq F Pr(>F)
1 10254 7800.6
2 10245 7736.8 9 63.799 9.3868 2.396e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Next, let’s compare if having a distinct intercept for each source improves the fit of the model.
Code
m_1_source <- lmer(log(frequency) ~ 1 + (1|source),
data = obese_annotated_filtered, REML = T)
m_1_sourceyear <- lmer(log(frequency) ~ scaled_year + (1|source),
data = obese_annotated_filtered, REML = T)
m_1_sourceyear2 <- lmer(log(frequency) ~ scaled_year + (scaled_year|source),
data = obese_annotated_filtered, REML = T)
m_1_sourceyeartype <- lmer(log(frequency) ~ scaled_year + source_type + (1|source),
data = obese_annotated_filtered, REML = T)
m_1_sourceyeartype2 <- lmer(log(frequency) ~ scaled_year + source_type + (scaled_year|source),
data = obese_annotated_filtered, REML = T)
rbind(
{broom::glance(m_0_sourceyear)%>%
dplyr::select(AIC, BIC, logLik) %>%
mutate(model = "scaled_year + source")},
{broom.mixed::glance(m_1_source) %>%
dplyr::select(AIC, BIC, logLik) %>%
mutate(model = "1 + (1/source)")},
{broom.mixed::glance(m_1_sourceyear)%>%
dplyr::select(AIC, BIC, logLik) %>%
mutate(model = "scaled_year + 1/source")},
{broom.mixed::glance(m_1_sourceyeartype)%>%
dplyr::select(AIC, BIC, logLik) %>%
mutate(model = "scaled_year + source_type + 1/source")},
{broom.mixed::glance(m_1_sourceyear2)%>%
dplyr::select(AIC, BIC, logLik) %>%
mutate(model = "scaled_year + scaled_year/source")},
{broom.mixed::glance(m_1_sourceyeartype2)%>%
dplyr::select(AIC, BIC, logLik) %>%
mutate(model = "scaled_year + source_type + scaled_year/source")}
) %>%
arrange(AIC) %>%
kable()| AIC | BIC | logLik | model |
|---|---|---|---|
| 26238.41 | 26325.24 | -13107.20 | scaled_year + source |
| 26239.73 | 26290.38 | -13112.86 | scaled_year + source_type + scaled_year/source |
| 26256.38 | 26299.79 | -13122.19 | scaled_year + scaled_year/source |
| 26275.94 | 26312.12 | -13132.97 | scaled_year + source_type + 1/source |
| 26292.53 | 26321.47 | -13142.26 | scaled_year + 1/source |
| 26296.20 | 26317.91 | -13145.10 | 1 + (1/source) |
Including a distinct intercept for each source did not drastically improve the fit of the model (AIC increases, logLik decreases; BIC does decrease slightly, so based on this criteria it may be a slightly better fit).
Let’s plot the residuals for the fixed effects model:
Code
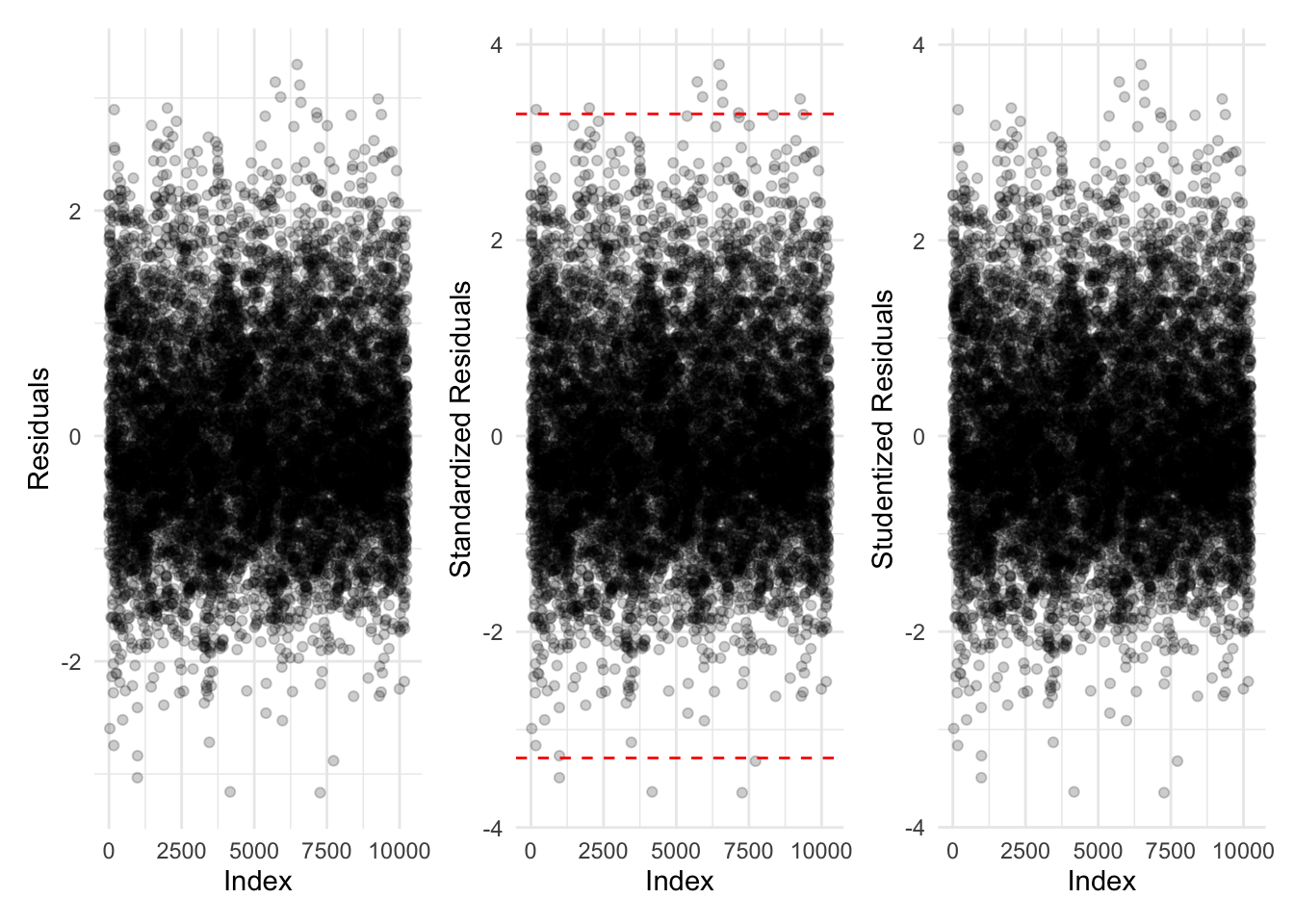
Proportion of data points with:
- abs(standardized residuals) > 3.29: 0.13%
- abs(standardized residuals) > 2.58: 1.02%
- abs(standardized residuals) > 1.96: 4.93%
All of these indicate the model is performing reasonably well on the data.
Let’s summarise the model (note that the intercept corresponds to the “first” source, i.e. the Advertiser):
Code
sjPlot::tab_model(m_0_sourceyear) | log(frequency) | |||
|---|---|---|---|
| Predictors | Estimates | CI | p |
| (Intercept) | 1.27 | 1.22 – 1.31 | <0.001 |
| source [Age] | -0.55 | -0.62 – -0.48 | <0.001 |
| source [Australian] | -0.63 | -0.71 – -0.55 | <0.001 |
| source [CanTimes] | -0.37 | -0.44 – -0.30 | <0.001 |
| source [CourierMail] | 0.01 | -0.06 – 0.07 | 0.816 |
| source [HeraldSun] | 0.02 | -0.05 – 0.08 | 0.628 |
| source [HobMercury] | 0.14 | 0.06 – 0.23 | 0.001 |
| source [NorthernT] | 0.23 | 0.13 – 0.33 | <0.001 |
| source [SydHerald] | -0.52 | -0.59 – -0.46 | <0.001 |
| source [WestAus] | 0.02 | -0.05 – 0.10 | 0.552 |
| scaled year | -0.01 | -0.02 – -0.01 | <0.001 |
| Observations | 10256 | ||
| R2 / R2 adjusted | 0.097 / 0.096 | ||
In summary, the fixed effects model which included source and year provided a good fit. Let’s summarise this model:
Code
report::report(m_0_sourceyear)We fitted a linear model (estimated using OLS) to predict frequency with source and scaled_year (formula: log(frequency) ~ source + scaled_year). The model explains a statistically significant and weak proportion of variance (R2 = 0.10, F(10, 10245) = 109.54, p < .001, adj. R2 = 0.10). The model’s intercept, corresponding to source = Advertiser and scaled_year = 0, is at 1.27 (95% CI [1.22, 1.31], t(10245) = 54.39, p < .001). Within this model:
- The effect of source [Age] is statistically significant and negative (beta = -0.55, 95% CI [-0.62, -0.48], t(10245) = -15.82, p < .001; Std. beta = -0.21, 95% CI [-0.23, -0.18])
- The effect of source [Australian] is statistically significant and negative (beta = -0.63, 95% CI [-0.71, -0.55], t(10245) = -15.68, p < .001; Std. beta = -0.23, 95% CI [-0.27, -0.20])
- The effect of source [CanTimes] is statistically significant and negative (beta = -0.37, 95% CI [-0.44, -0.30], t(10245) = -9.90, p < .001; Std. beta = -0.16, 95% CI [-0.19, -0.13])
- The effect of source [CourierMail] is statistically non-significant and positive (beta = 7.70e-03, 95% CI [-0.06, 0.07], t(10245) = 0.23, p = 0.816; Std. beta = -6.00e-03, 95% CI [-0.03, 0.02])
- The effect of source [HeraldSun] is statistically non-significant and positive (beta = 0.02, 95% CI [-0.05, 0.08], t(10245) = 0.48, p = 0.628; Std. beta = -8.24e-03, 95% CI [-0.03, 0.02])
- The effect of source [HobMercury] is statistically significant and positive (beta = 0.14, 95% CI [0.06, 0.23], t(10245) = 3.31, p < .001; Std. beta = 0.06, 95% CI [0.02, 0.09])
- The effect of source [NorthernT] is statistically significant and positive (beta = 0.23, 95% CI [0.13, 0.33], t(10245) = 4.44, p < .001; Std. beta = 0.07, 95% CI [0.03, 0.11])
- The effect of source [SydHerald] is statistically significant and negative (beta = -0.52, 95% CI [-0.59, -0.46], t(10245) = -16.02, p < .001; Std. beta = -0.21, 95% CI [-0.23, -0.18])
- The effect of source [WestAus] is statistically non-significant and positive (beta = 0.02, 95% CI [-0.05, 0.10], t(10245) = 0.59, p = 0.552; Std. beta = -0.01, 95% CI [-0.04, 0.02])
- The effect of scaled year is statistically significant and negative (beta = -0.01, 95% CI [-0.02, -5.16e-03], t(10245) = -3.96, p < .001; Std. beta = -0.02, 95% CI [-0.02, -8.05e-03])
Standardized parameters were obtained by fitting the model on a standardized version of the dataset. 95% Confidence Intervals (CIs) and p-values were computed using the Wald approximation.
To summarise, a model was fit by source and year, which explained a small amount of variance in the data. It showed that the Age, Australian, Canberra Times and Sydney Morning Herald had a lower frequency of use of the adjective lemma OBESE relative to the Advertiser, while in the Hobart Mercury, Northern Territorian the adjective lemma OBESE was used somewhat more frequently than in the Advertiser. Use of the adjective lemma OBESE decreased with time in the corpus.
If we look at the mixed effects model scaled_year + source_type + scaled_year/source, which had the lowest BIC, we can see that the observation of higher frequency of counts in tabloids is reproduced, while the signal from the decrease by year is not detected:
Code
sjPlot::tab_model(m_1_sourceyeartype2)| log(frequency) | |||
|---|---|---|---|
| Predictors | Estimates | CI | p |
| (Intercept) | 0.75 | 0.65 – 0.84 | <0.001 |
| scaled year | -0.01 | -0.02 – 0.00 | 0.054 |
| source type [tabloid] | 0.58 | 0.46 – 0.70 | <0.001 |
| Random Effects | |||
| σ2 | 0.75 | ||
| τ00source | 0.01 | ||
| τ11source.scaled_year | 0.00 | ||
| ρ01source | -0.18 | ||
| ICC | 0.02 | ||
| N source | 10 | ||
| Observations | 10256 | ||
| Marginal R2 / Conditional R2 | 0.100 / 0.114 | ||
The intercepts and slopes for scaled year for each of the sources were:
| (Intercept) | scaled_year | source_typetabloid | |
|---|---|---|---|
| Australian | 0.6482964 | -0.0153703 | 0.5831533 |
| CourierMail | 0.6838377 | 0.0273333 | 0.5831533 |
| Advertiser | 0.6941022 | -0.0378463 | 0.5831533 |
| HeraldSun | 0.7009579 | -0.0077894 | 0.5831533 |
| WestAus | 0.7057935 | -0.0152603 | 0.5831533 |
| Age | 0.7172206 | -0.0107049 | 0.5831533 |
| SydHerald | 0.7434942 | -0.0139594 | 0.5831533 |
| HobMercury | 0.8135840 | -0.0111710 | 0.5831533 |
| NorthernT | 0.8725238 | -0.0212058 | 0.5831533 |
| CanTimes | 0.8825098 | -0.0163459 | 0.5831533 |
Differences in usage by topic
Code
| topic_label | broadsheet | tabloid | total_topic | prop_broadsheet |
|---|---|---|---|---|
| ChildrenParents | 364 | 1335 | 1699 | 21.42 |
| NutritionStudy | 642 | 1797 | 2439 | 26.32 |
| WomenPregnancy | 159 | 385 | 544 | 29.23 |
| FitnessExercise | 464 | 1093 | 1557 | 29.80 |
| BiomedResearch | 1013 | 2077 | 3090 | 32.78 |
| MedicalHealth | 297 | 524 | 821 | 36.18 |
| Food | 539 | 800 | 1339 | 40.25 |
| FastFood&Drinks | 1044 | 1441 | 2485 | 42.01 |
| Transport&Commuting | 427 | 559 | 986 | 43.31 |
| WomenGirls | 625 | 744 | 1369 | 45.65 |
| Awards | 28 | 33 | 61 | 45.90 |
| PublicHealthReport | 1243 | 1338 | 2581 | 48.16 |
| Students&Teachers | 536 | 572 | 1108 | 48.38 |
| Microbiome | 32 | 33 | 65 | 49.23 |
| SportsDoping | 260 | 253 | 513 | 50.68 |
| Politics | 1478 | 1249 | 2727 | 54.20 |
| MusicMovies | 1543 | 1236 | 2779 | 55.52 |
We can see that articles labelled with the topics “ChildrenParents”, “NutritionStudy”, “WomenPregnancy” and “FitnessExercise” are approximately 3x more frequently reported on in tabloids than broadsheets, in contrast to topics like “Politics” and “SportsDoping” which are approximately evenly represented between the two media types.
Code
obese_annotated_filtered %>%
ggplot(aes(x = as.factor(
reorder(topic_label,frequency)),
y = log(frequency),
fill = topic_label)) +
geom_boxplot() +
theme(axis.text.x =
element_text(angle = 90, vjust = 0.5, hjust=1),
legend.position = "NA") +
labs(x = "Topic label",
y = "log(frequency per 1000 words)")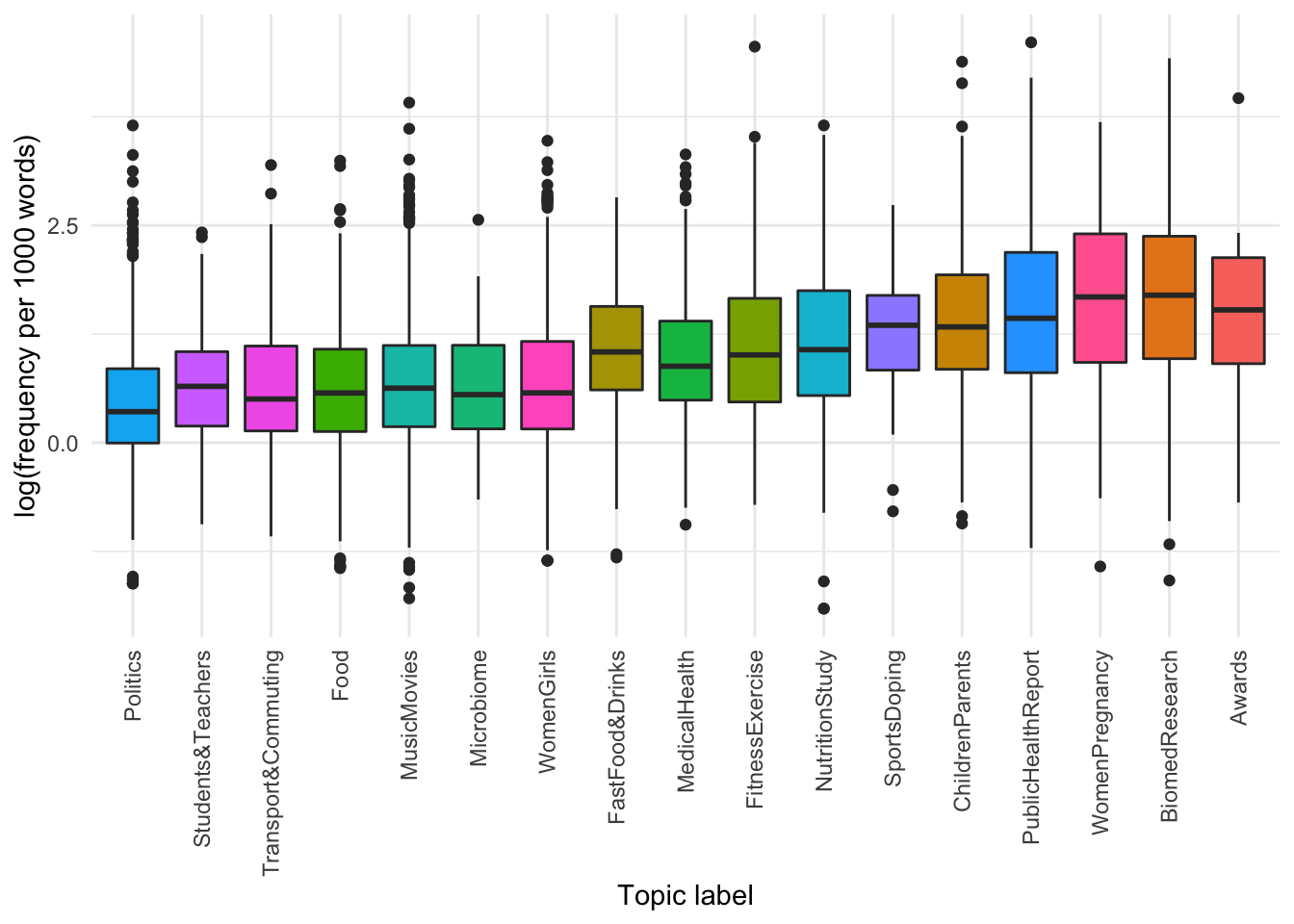
It seems there are some topics that use OBESE more than others.
We use a simple linear model with post-hoc comparisons and Bonferroni multiple testing correction:
Code
obese_by_topic <- lm(
frequency ~ as.factor(topic_label),
data = obese_annotated_filtered)
library(emmeans)
obese_by_topic_comp <- emmeans(obese_by_topic, pairwise ~ as.factor(topic_label), adjust = "bonferroni")
obese_by_topic_comp$contrasts %>%
summary(infer = TRUE) %>%
filter(p.value < 0.01) %>%
select(-df) |>
kable()| contrast | estimate | SE | lower.CL | upper.CL | t.ratio | p.value |
|---|---|---|---|---|---|---|
| Awards - Food | 5.942155 | 1.4853012 | 0.6493259 | 11.2349838 | 4.000640 | 0.0086518 |
| Awards - Politics | 6.288435 | 1.4729512 | 1.0396145 | 11.5372546 | 4.269276 | 0.0026911 |
| Awards - Students&Teachers | 6.172930 | 1.4968448 | 0.8389660 | 11.5068943 | 4.123961 | 0.0051048 |
| Awards - Transport&Commuting | 5.942849 | 1.4971190 | 0.6079082 | 11.2777906 | 3.969524 | 0.0098612 |
| BiomedResearch - ChildrenParents | 2.635277 | 0.2323342 | 1.8073605 | 3.4631932 | 11.342613 | 0.0000000 |
| BiomedResearch - FastFood&Drinks | 4.493757 | 0.2333711 | 3.6621454 | 5.3253683 | 19.255838 | 0.0000000 |
| BiomedResearch - FitnessExercise | 3.872260 | 0.2219288 | 3.0814229 | 4.6630966 | 17.448211 | 0.0000000 |
| BiomedResearch - Food | 5.623753 | 0.2991564 | 4.5577179 | 6.6897886 | 18.798706 | 0.0000000 |
| BiomedResearch - MedicalHealth | 4.461614 | 0.3299740 | 3.2857608 | 5.6374669 | 13.521107 | 0.0000000 |
| BiomedResearch - Microbiome | 5.446934 | 1.1570708 | 1.3237445 | 9.5701229 | 4.707520 | 0.0003455 |
| BiomedResearch - MusicMovies | 5.496256 | 0.1920506 | 4.8118893 | 6.1806227 | 28.618798 | 0.0000000 |
| BiomedResearch - NutritionStudy | 3.717134 | 0.2237552 | 2.9197880 | 4.5144790 | 16.612498 | 0.0000000 |
| BiomedResearch - Politics | 5.970033 | 0.2301306 | 5.1499692 | 6.7900968 | 25.941938 | 0.0000000 |
| BiomedResearch - PublicHealthReport | 1.510443 | 0.2134401 | 0.7498552 | 2.2710307 | 7.076659 | 0.0000000 |
| BiomedResearch - Students&Teachers | 5.854529 | 0.3520215 | 4.6001098 | 7.1089473 | 16.631166 | 0.0000000 |
| BiomedResearch - Transport&Commuting | 5.624448 | 0.3531856 | 4.3658810 | 6.8830145 | 15.924909 | 0.0000000 |
| BiomedResearch - WomenGirls | 5.197677 | 0.2446344 | 4.3259286 | 6.0694243 | 21.246710 | 0.0000000 |
| ChildrenParents - FastFood&Drinks | 1.858480 | 0.2705723 | 0.8943035 | 2.8226567 | 6.868701 | 0.0000000 |
| ChildrenParents - FitnessExercise | 1.236983 | 0.2607674 | 0.3077457 | 2.1662201 | 4.743626 | 0.0002894 |
| ChildrenParents - Food | 2.988476 | 0.3290012 | 1.8160898 | 4.1608630 | 9.083481 | 0.0000000 |
| ChildrenParents - MedicalHealth | 1.826337 | 0.3572536 | 0.5532740 | 3.0994000 | 5.112159 | 0.0000441 |
| ChildrenParents - MusicMovies | 2.860979 | 0.2358616 | 2.0204932 | 3.7014651 | 12.129909 | 0.0000000 |
| ChildrenParents - NutritionStudy | 1.081857 | 0.2623236 | 0.1470739 | 2.0166394 | 4.124130 | 0.0051011 |
| ChildrenParents - Politics | 3.334756 | 0.2677823 | 2.3805216 | 4.2889907 | 12.453238 | 0.0000000 |
| ChildrenParents - PublicHealthReport | -1.124834 | 0.2535822 | -2.0284669 | -0.2212008 | -4.435776 | 0.0012607 |
| ChildrenParents - Students&Teachers | 3.219252 | 0.3777121 | 1.8732854 | 4.5652181 | 8.523031 | 0.0000000 |
| ChildrenParents - Transport&Commuting | 2.989171 | 0.3787972 | 1.6393379 | 4.3390039 | 7.891218 | 0.0000000 |
| ChildrenParents - WomenGirls | 2.562400 | 0.2803449 | 1.5633986 | 3.5614007 | 9.140169 | 0.0000000 |
| ChildrenParents - WomenPregnancy | -2.125124 | 0.3656700 | -3.4281787 | -0.8220691 | -5.811589 | 0.0000009 |
| FastFood&Drinks - MusicMovies | 1.002499 | 0.2368831 | 0.1583730 | 1.8466252 | 4.232042 | 0.0031769 |
| FastFood&Drinks - Politics | 1.476276 | 0.2686824 | 0.5188338 | 2.4337183 | 5.494502 | 0.0000055 |
| FastFood&Drinks - PublicHealthReport | -2.983314 | 0.2545326 | -3.8903337 | -2.0762942 | -11.720753 | 0.0000000 |
| FastFood&Drinks - WomenPregnancy | -3.983604 | 0.3663297 | -5.2890096 | -2.6781982 | -10.874367 | 0.0000000 |
| FitnessExercise - Food | 1.751494 | 0.3217375 | 0.6049911 | 2.8979959 | 5.443859 | 0.0000073 |
| FitnessExercise - MusicMovies | 1.623996 | 0.2256189 | 0.8200098 | 2.4279827 | 7.197963 | 0.0000000 |
| FitnessExercise - Politics | 2.097773 | 0.2588060 | 1.1755254 | 3.0200210 | 8.105582 | 0.0000000 |
| FitnessExercise - PublicHealthReport | -2.361817 | 0.2440843 | -3.2316042 | -1.4920294 | -9.676235 | 0.0000000 |
| FitnessExercise - Students&Teachers | 1.982269 | 0.3714022 | 0.6587875 | 3.3057501 | 5.337256 | 0.0000131 |
| FitnessExercise - Transport&Commuting | 1.752188 | 0.3725057 | 0.4247745 | 3.0796015 | 4.703789 | 0.0003519 |
| FitnessExercise - WomenGirls | 1.325417 | 0.2717838 | 0.3569227 | 2.2939107 | 4.876731 | 0.0001489 |
| FitnessExercise - WomenPregnancy | -3.362107 | 0.3591487 | -4.6419229 | -2.0822907 | -9.361323 | 0.0000000 |
| Food - NutritionStudy | -1.906620 | 0.3230001 | -3.0576213 | -0.7556181 | -5.902846 | 0.0000005 |
| Food - PublicHealthReport | -4.113310 | 0.3159419 | -5.2391605 | -2.9874601 | -13.019197 | 0.0000000 |
| Food - WomenPregnancy | -5.113600 | 0.4113757 | -6.5795258 | -3.6476747 | -12.430488 | 0.0000000 |
| MedicalHealth - Politics | 1.508419 | 0.3558244 | 0.2404489 | 2.7763894 | 4.239223 | 0.0030772 |
| MedicalHealth - PublicHealthReport | -2.951171 | 0.3452646 | -4.1815114 | -1.7208304 | -8.547564 | 0.0000000 |
| MedicalHealth - WomenPregnancy | -3.951461 | 0.4343020 | -5.4990839 | -2.4038379 | -9.098417 | 0.0000000 |
| Microbiome - WomenPregnancy | -4.936781 | 1.1910282 | -9.1809762 | -0.6925853 | -4.144974 | 0.0046592 |
| MusicMovies - NutritionStudy | -1.779122 | 0.2274157 | -2.5895120 | -0.9687330 | -7.823217 | 0.0000000 |
| MusicMovies - PublicHealthReport | -3.985813 | 0.2172744 | -4.7600643 | -3.2115618 | -18.344603 | 0.0000000 |
| MusicMovies - WomenPregnancy | -4.986103 | 0.3414950 | -6.2030107 | -3.7691954 | -14.600810 | 0.0000000 |
| NutritionStudy - Politics | 2.252899 | 0.2603739 | 1.3250644 | 3.1807345 | 8.652555 | 0.0000000 |
| NutritionStudy - PublicHealthReport | -2.206691 | 0.2457462 | -3.0824000 | -1.3309811 | -8.979553 | 0.0000000 |
| NutritionStudy - Students&Teachers | 2.137395 | 0.3724965 | 0.8100143 | 3.4647759 | 5.738027 | 0.0000013 |
| NutritionStudy - Transport&Commuting | 1.907314 | 0.3735967 | 0.5760128 | 3.2386157 | 5.105275 | 0.0000457 |
| NutritionStudy - WomenGirls | 1.480543 | 0.2732773 | 0.5067270 | 2.4543590 | 5.417731 | 0.0000084 |
| NutritionStudy - WomenPregnancy | -3.206980 | 0.3602802 | -4.4908287 | -1.9231323 | -8.901352 | 0.0000000 |
| Politics - PublicHealthReport | -4.459590 | 0.2515648 | -5.3560340 | -3.5631460 | -17.727402 | 0.0000000 |
| Politics - WomenPregnancy | -5.459880 | 0.3642739 | -6.7579598 | -4.1618003 | -14.988392 | 0.0000000 |
| PublicHealthReport - Students&Teachers | 4.344086 | 0.3663931 | 3.0384541 | 5.6497171 | 11.856351 | 0.0000000 |
| PublicHealthReport - Transport&Commuting | 4.114005 | 0.3675116 | 2.8043875 | 5.4236221 | 11.194217 | 0.0000000 |
| PublicHealthReport - WomenGirls | 3.687234 | 0.2648976 | 2.7432783 | 4.6311888 | 13.919465 | 0.0000000 |
| Students&Teachers - WomenPregnancy | -5.344376 | 0.4512810 | -6.9525027 | -3.7362485 | -11.842678 | 0.0000000 |
| Transport&Commuting - WomenPregnancy | -5.114295 | 0.4521896 | -6.7256596 | -3.5029300 | -11.310067 | 0.0000000 |
| WomenGirls - WomenPregnancy | -4.687524 | 0.3736059 | -6.0188577 | -3.3561893 | -12.546705 | 0.0000000 |
We can see that there are differences in the frequency of use of the adjective lemma OBESE by topic, with articles annotated as “Awards” and “Bio-medical Research” using more instances per 1000 words than articles discussing politics, schooling, transport and commuting.