Automating with Arrays Jobs
Overview
Teaching: 25 min
Exercises: 5 minQuestions
Using array jobs to ‘batch’ analyses
Objectives
Explore using job arrays to batch multiple jobs
Get to know the
awkcommand
Automating with Arrays Jobs
This episode introduces different ways to use array jobs to automate, or ‘batch’, computations on Artemis.
Using the Array Index as a parameter
The array index takes integer values in a sequence, as defined with the -J i-f:s PBS option. In some cases, you may be able to directly use these indices to refer to the specific input files, directories or parameters you want as inputs for your array of compute jobs. For example:
-
You might have input data files named inputDataN.dat, which you could invoke in your PBS script as
inputData${PBS_ARRAY_INDEX}.dat -
You might have data from different subjects in numbered folders subjectN, invoked as
data/subject$PBS_ARRAY_INDEX -
You might have a set of parameters you wish to use; eg you could test rates from 0.1 to 1.0 with
rate=$(echo "scale=2; $PBS_ARRAY_INDEX/10" | bc), using the POSIX ‘basic calculator’ functionbcto divide by 10
The next example exercise uses this last method to test different length substrings (k-mers) of a DNA sequence. Navigate to the Assembly folder in the data we extracted earlier, and open the assembly.pbs script file:
cd ../Assembly
nano assembly.pbs
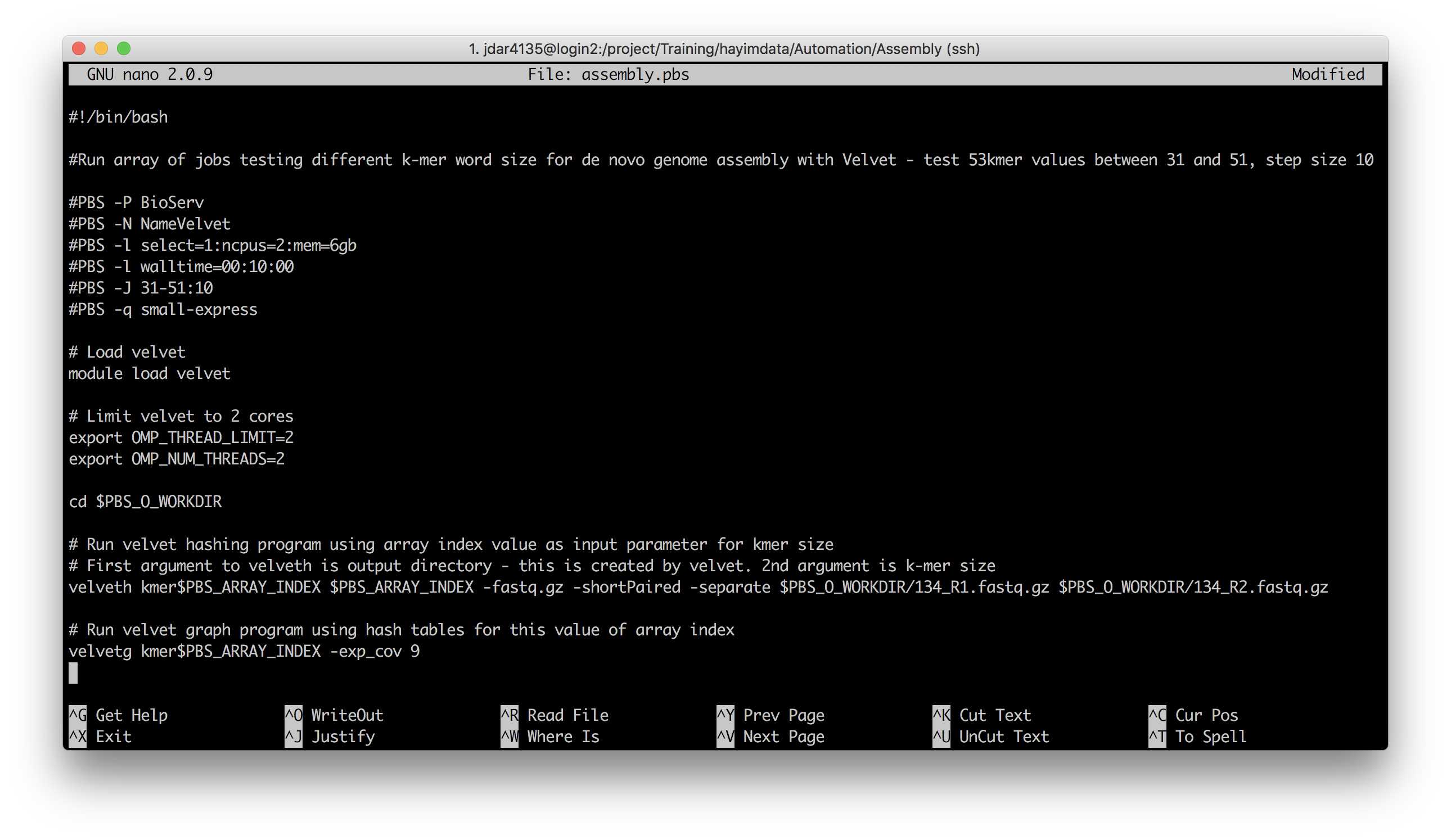
This script tests k-mer lengths from 31 to 51 in steps of 10 – so 31, 41 and 51. Since these are integer values, no fancy math operations are needed, just the array index itself.
The 2nd argument to the velveth function called in this script is k-mer word length. The function also takes a directory name as its 1st argument, and outputs the results into there.
Make any changes to the script as required, and then submit it with qsub.
Change #1
Specify your project.
Use the
-PPBS directive to specify the Training project, using its short name.#PBS -P Training
Change #2
Give your job a name
Use the
-NPBS directive to give your job an easily identifiable name. You might run lots of jobs at the same time, so you want to be able to keep track of them!#PBS -N VelvetHayimSubstitute a job name of your choice!
Change #3
If you are using the Training Scheduler1 then you do not have access to all the queues. You can submit jobs to defaultQ and dtq only.
In the normal Artemis environment you can submit to defaultQ, dtq, small-express, scavenger, and possibly some strategic allocation queues you may have access to.
#PBS -q defaultQ
What two Bash variables are referenced in this script, and what two are set?
Answer
Referenced:
(these variables are set by PBS)
PBS_O_WORKDIR
PBS_ARRAY_INDEXSet:
(these variables will be queried by velveth)
OMP_THREAD_LIMIT
OMP_NUM_THREADS
How might an additional PBS-initialised Bash variable be used in this script?
Answer
The number of CPUs to use could be automatically set to the number requested for the job, with
NCPUS:# Limit velvet to 2 cores export OMP_THREAD_LIMIT=$NCPUS export OMP_NUM_THREADS=$NCPUS
Monitor your job’s status with qstat -t. When all subjobs have completed, check for any non-zero exit statuses, indicating errors:
grep -L "Exit Status: 0" VelvetHayim*usage | xargs cat
The above set of commands looks for _usage log files which do not contain “Exit Status: 0”, and then calls xargs to send them to the cat function, printing their contents to the terminal output. Make sure to use your job’s name, not mine!
Using the Array Index with a config file
Configuration files
Sometimes you may wish to change more than one value at a time. For example, you might have a whole set of similar analyses to run, each with its own inputs or settings, for which you would need to write a separate PBS script each.
This problem can be solved with a configuration (‘config’) file. A config file is basically a list of settings and parameters for your data, with one analysis per row, and each column a different setting. (Obviously, you can arrange this information however you want, but this is simplest!)
Navigate to the Alignment folder, and have a look at the file samples.config:
cd ../Alignment
cat samples.config
[jdar4135@login3 Alignment]$ cat samples.config
#ArrayIndex SampleID Breed Reference SeqCentre RunIDs
1 USCF70 Dalmatian canfam3_chr20.fasta Ramaciotti D09NUACXX
2 BD394 Boxer canfam3_chr20.fasta UCDavis C7RNWACXX,C16NWHCXX
3 FM0238 FranchesM equcab2_chr20.fasta UBern D1A03ACXX,C176WACXX,D16LHACXX
4 FM0570 FranchesM equcab2_chr20.fasta UBern D16LHACXX,D1A03ACXX
samples.config demonstrates an example configuration file. The first line (a comment) lists the column headers. Each line after defines an analysis to be run. There are four analyses (1-4), each defined by their input data (.fastq raw data and .fasta reference files), and various metadata. The aim is to write a single PBS script that will run all 4.
Parsing with awk
Awk is one of those very powerful, somewhat intimidating Linux tools2 that is well worth learning. The awk command operates over text data line-by-line, performing operations over specified columns. Extra steps can also be added before and after all the lines of data have been read, which can be handy for computing summary statistics over a list of values.
A typical usage of the awk command might look like:
breed=$(awk -v taskID=$PBS_ARRAY_INDEX '$1==taskID {print $3}' $config)
In the above command, awk is called inside a Bash expansion call $(...)– the result of what is evaluated inside the parentheses is then stored in a variable called breed.
Let’s consider the awk command itself:
awk -v taskID=$PBS_ARRAY_INDEX '$1==taskID {print $3}' $config
The first term calls awk itself. The main argument to awk is what follows inside the '' apostrophes. Before that, there is a -v argument, which is used to pass variables from your current Bash session into awk’s memory space; in this case, the PBS_ARRAY_INDEX variable of the subjob is passed to awk under the name ‘taskID’.
The last argument, $config points to the input text we want awk to operate over; this variable was set to point to the samples.config file earlier in the script.
The second argument to awk contains the operations we want awk to run, listed between '' single apostrophes. Numbered variables, eg $1 and $3 refer to the columns of the text input, with the leftmost column being 1. Any relational operators (==, or >, etc) are treated as conditionals; any commands relating to a conditional follow it inside {}.
So, the command above tells awk to find where column 1 is equal to taskID, and then print out the contents of column 3. Eg, for subjob #2 (when taskID = $PBS_ARRAY_INDEX is 2) this would find the 2nd data row in samples.config:
2 BD394 Boxer canfam3_chr20.fasta UCDavis C7RNWACXX,C16NWHCXX
and print out the ‘Breed’ entry (column 3). This output is ‘printed’ to stdout, but since the whole command was enclosed in an expansion and assignment breed=$(..), this result, ‘Boxer’, is stored in the breed variable in the script.
Take a minute to process all this.. when you think you have it, let’s take a look at the whole PBS script.
Reading in job parameters from a config file
Open align.pbs in your preferred text editor.
nano align.pbs
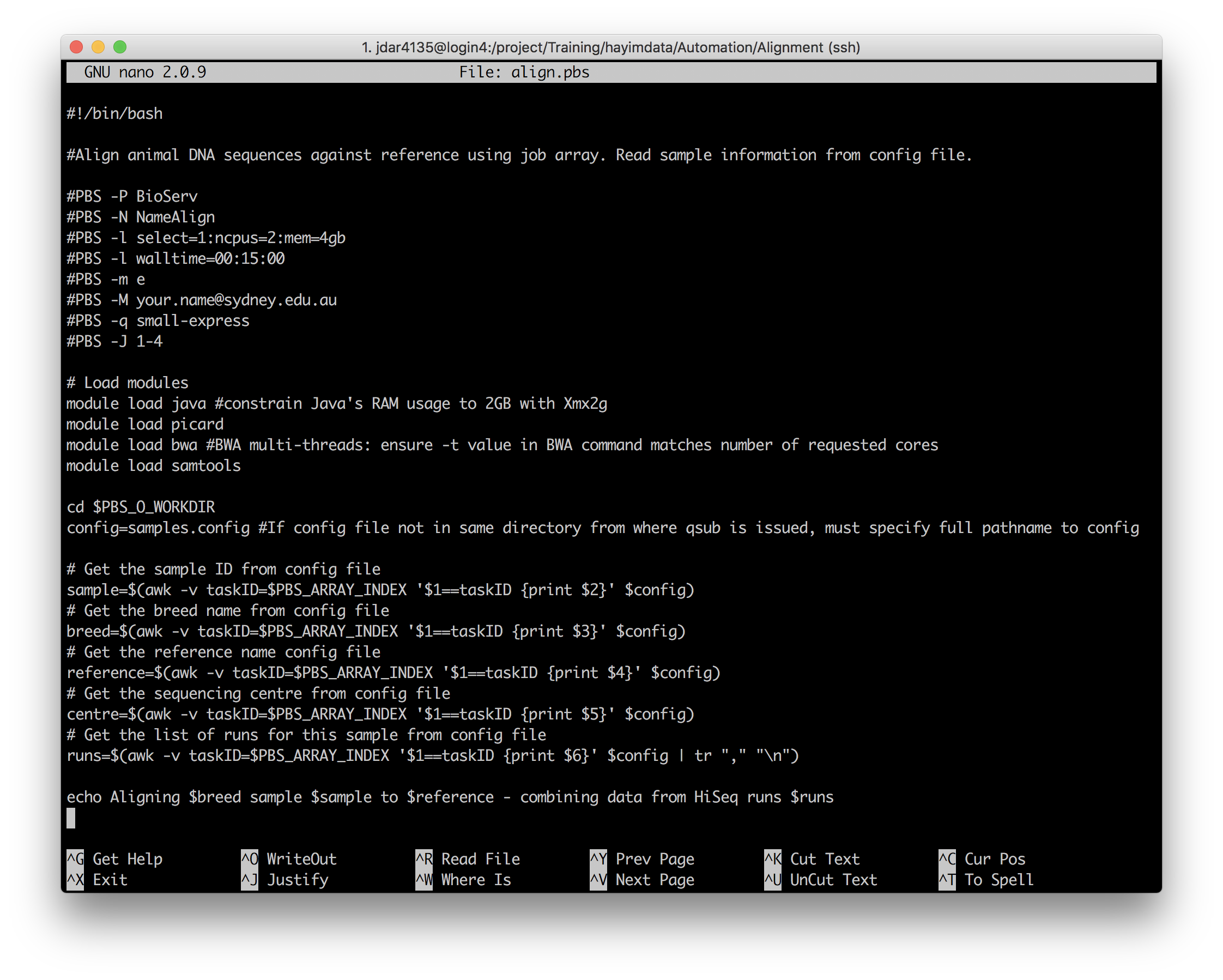
Have a look at the lines from config= onwards.
First, the input configuration file, samples.config, is declared into a variable, so it won’t have to be written out multiple times.
Then, there are 5 calls to awk, each enclosed by an expansion and assignment to a Bash variable foo=$(bar): sample, breed, reference, centre, and runs. Each of these commands uses awk to extract the data from one of the columns in samples.config and store it in a variable. The row that the data is taken from is determined by the array index of the subjob – the conditional $1=taskID matches the $PBS_ARRAY_INDEX number of the running subjob to the indices in the first column of the config data.
Make any required changes to the PBS script, and submit it with qsub. Then, have a look at the rest of the script to see how it uses the config data it has just read in.
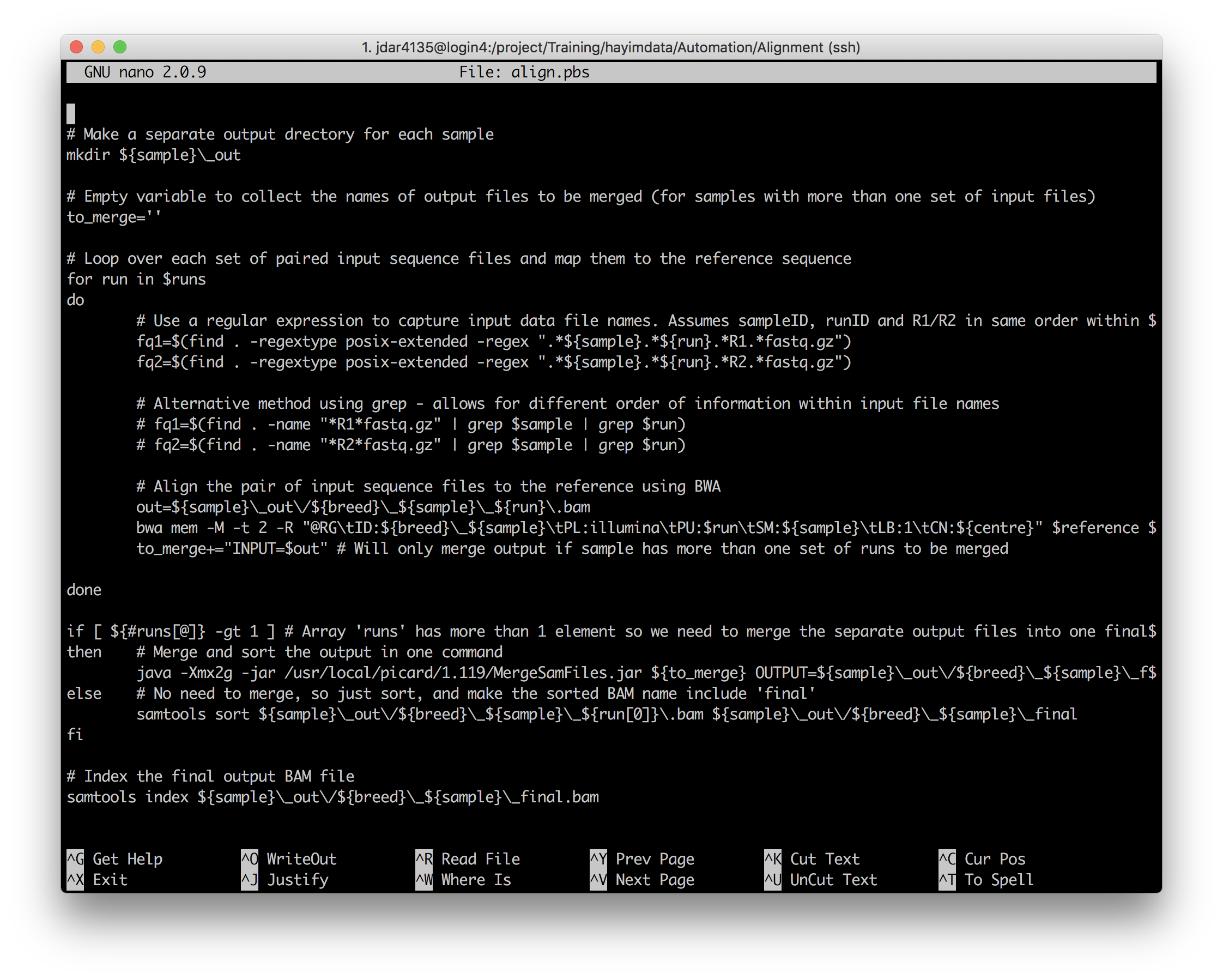
Which variable is only used once per run?
Answer
centreis only used once! On line 55.
Monitor your jobs with qstat -t, and then check for output and log files to check the array ran correctly. You should have a new output directory for each sample, containing some BAM and BAI files.
While we’re waiting for these jobs to finish, let’s continue to the next Episode.
Notes
1↩As you should recall from the Introduction to Artemis HPC course, the scheduler is the software that runs the cluster, allocating jobs to physical compute resources. Artemis HPC provides us with a separate ‘mini-cluster’ for Training, which has a separate PBS scheduler instance and dedicated resources.
2↩In fact, Awk is really a shell, a programming language and a command all in one!
Key Points
The
PBS_ARRAY_INDEXcan be used to index input data
awkis your friend!