Optimisation
Overview
Teaching: 15 min
Exercises: 10 minQuestions
Selecting the appropriate resources for your job to maximise efficiency
Objectives
Understand optimisation and CPU efficiency
Learn how to use benchmarking and usage statistics to select optimal resources for a job
Optimisation
In order to make the best use of your research time and KSU, a little time spent at the outset of a new analysis performing benchmarking and optimisation is worthwhile.
NCI regularly monitors job performance, and jobs with consistently low CPU efficiency are identified and their users contacted to troubleshoot.
CPU efficiency
This refers to how well the CPU requested for the job are being utilised, and is commonly expressed as a percentage or range from 0 - 1. It is calculated as CPUtime / Walltime / CPUs.
While 100% is optimal, most jobs don’t achieve this, particularly those that use multi-threaded applcations (where there is usually a decline in CPU efficiency with increasing threads), those with heavy I/O, and jobs where multiple steps are chained together. Generally we aim for a minimum of 70% CPU efficiency.
For workflows with multiple steps chained together, benchmarking the optimum resources for each individual command in the job can help identify which commands should be included in separate jobs. Breaking a complex workflow into multiple jobs has the added benefit of creating natural checkpoints, making it easier to rerun failed components.
For multi-threading applications, benchmarking should be run at a range of threads on a representative subset of your input data to pick the optimal trade-off between walltime and CPU efficiency.
This script can be used to report usage statistics of completed Gadi jobs. You can also use the command nqstat_anu to observe the CPU efficiency of running jobs, or the qps command for each individual process.
Efficiency and scalability
In the next section, one of the parallel job examples uses the software BWA to map DNA sequence to a reference DNA database. Typically this is done for tens of millions of DNA strings on one node in one long-running job, but we have optimised this by physically splitting the very large input DNA file into lots of smaller files for parallel mapping. The initial benchmarking needs to answer two questions:
- Efficiency: What are the optimal number of CPUs to assign multi-threaded BWA for input files of this size
- Scalability: Will the CPU efficiency achieved for one such task at the chosen CPUs remain ~ constant when executed in parallel?
Efficiency and scalability are two critical aspects required for Gadi workflows. The NCMAS review panel rigorously scrutinises the computational part of applications to ensure these criteria are met. The SIH HPC Allocation panel similarly expects applications to demonstrate these attrributes, however the earleir rounds of the scheme have been somewhat lenient in order to encourage more Artemis users onto Gadi. Both SIH and NCI can assist you with optimising your workflows for Gadi, so please contact if you need assistance.
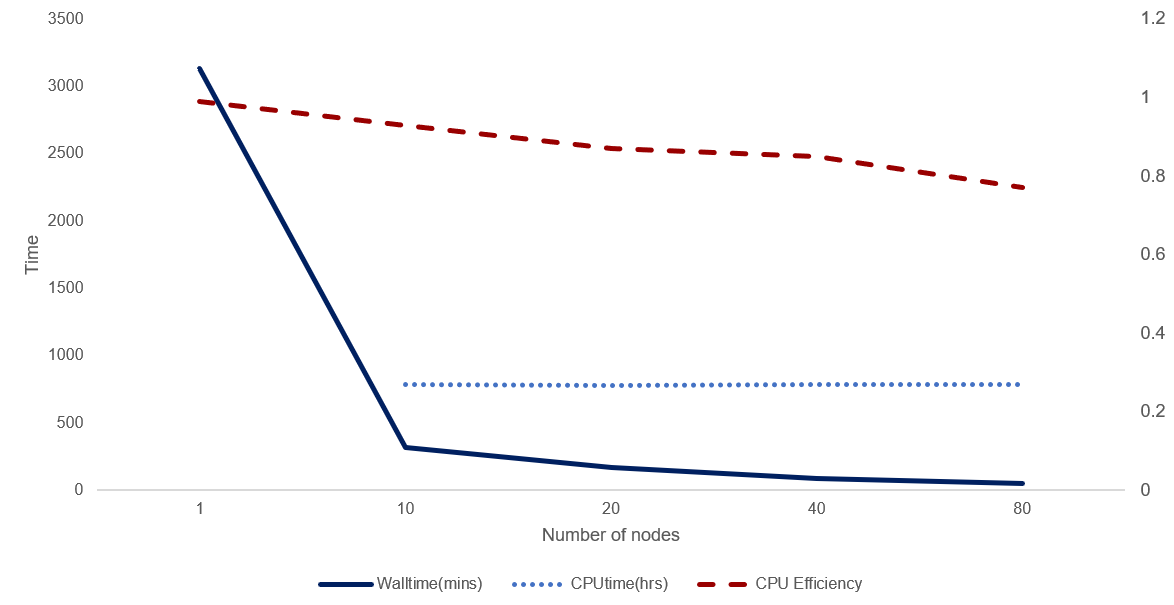
For a job to be considered efficient and scalable, the plot of walltime versus nodes should linearly decrease, ie for a given set of inputs, if double the nodes are used, the walltime will halve. For a perfect correlation like this, the CPU efficiency remains constant (horizontal). In practice the trend often declines, as CPU efficiency may decline with increasing CPUs/nodes.
Benchmarking example
Going back to our DNA example, we will now submit a handful of benchmarking jobs to help optimise our parallel mapping job.
If you missed the step about downloading sample data from Section 5 Data transfer to your Gadi scratch space, please do so now.
Unpack the download and change into the extracted directory (either Sample_data_benchmark or Sample_data_bio depending on which dataset you downloaded):
tar -zxvf Sample_data_<dataset>.tar.gz
cd Sample_data_<dataset>
We will submit ./Scripts/align_benchmark.pbs for five CPU values: 2, 4, 6, 8 and 12. When seleting the number of CPUs per parallel task, ensure that the selected values are divisible by the number of CPUs on the node. We will use Gadi’s ‘normal’ queue (or ‘express’ if you wish) - these have 48 CPUs per node. Others have 28 (Broadwell nodes).
Update the project code at -P and lstorage directives, then submit using a loop:
for NCPUS in 2 4 6 8 12
do
mem=$(expr $NCPUS \* 4)
qsub -N BA-${NCPUS} -l ncpus=${NCPUS},mem=${mem}GB -o ./Logs/benchmark_align_${NCPUS}CPUs.o -e ./Logs/benchmark_align_${NCPUS}CPUs.e -v NCPUS="${NCPUS}" Scripts/align_benchmark.pbs
sleep 2
done
![]() Note: ‘for’ loops for job submission are not recommended on Gadi! Their use here is purely for simplicity of the training exercise
Note: ‘for’ loops for job submission are not recommended on Gadi! Their use here is purely for simplicity of the training exercise
While the jobs are running (they are quick, so do this right away!) try out some of Gadi’s job monitoring commands:
nqstat_anu
qps <jobID>
Once all five jobs have successfully completed, you can generate TSV usage metrics by changing into the ./Logs direcctory, then running the following command:
cd Logs
perl ../Scripts/gadi_usage_report.pl benchmark_align
TSV will be printed to STDOUT unless redirected. Note this script is part of the SIH HPC usage reports repo, which also includes Artemis scripts, but has been copied here for convenience.
| #JobName | CPUs_requested | CPUs_used | Mem_requested | Mem_used | CPUtime | CPUtime_mins | Walltime_req | Walltime_used | Walltime_mins | JobFS_req | JobFS_used | Efficiency | Service_units |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| benchmark_align_2CPUs | 2 | 2 | 8.0GB | 967.17MB | 0:07:09 | 7.15 | 0:30:00 | 0:04:00 | 4 | 100.0MB | 0B | 0.89 | 0.27 |
| benchmark_align_4CPUs | 4 | 4 | 16.0GB | 1.17GB | 0:08:02 | 8.03 | 0:30:00 | 0:02:37 | 2.62 | 100.0MB | 0B | 0.77 | 0.35 |
| benchmark_align_6CPUs | 6 | 6 | 24.0GB | 1.68GB | 0:06:46 | 6.77 | 0:30:00 | 0:02:08 | 2.13 | 100.0MB | 0B | 0.53 | 0.43 |
| benchmark_align_8CPUs | 8 | 8 | 32.0GB | 1.91GB | 0:06:41 | 6.68 | 0:30:00 | 0:01:23 | 1.38 | 100.0MB | 0B | 0.61 | 0.37 |
| benchmark_align_12CPUs | 12 | 12 | 48.0GB | 2.2GB | 0:06:17 | 6.28 | 0:30:00 | 0:01:06 | 1.1 | 100.0MB | 0B | 0.48 | 0.44 |
![]() Looking at the efficiency and walltime, how many CPUs would you assign per parallel task?
Looking at the efficiency and walltime, how many CPUs would you assign per parallel task?
I would select 4: while this has slightly worse efficiency than 2, the saving in walltime will be valuable when scaling this up to a large dataset of say 100 whole genomes, where > 100,000 tasks would be expected.
![]() If your project was running low on KSU, you might choose 2, as the increased CPU efficiency provides a lower SU cost per task.
If your project was running low on KSU, you might choose 2, as the increased CPU efficiency provides a lower SU cost per task.
Note that the CPU time declines as CPUs per task increases. This suggests that the total amount of work for the job is decreasing as more CPUs are assigned to each task. You might explore this for your code/software, to see if you can try and optimise so that as much CPU time is used as possible for the wallime of the job. It is the walltime that is used when calculating the KSU charge for the job, so if you can make the CPU work harder during that walltime, your job will be more efficient and cost less.
Scalability tests
Before executing parallel jobs, it is beneficial to run some saclability tests after your benchmarking, eg if your parallel job will have 100,000 tasks, run subset jobs of say 100 and 1,000 tasks first: does the CPU efficiency remain stable as nodes and simultaneous tasks increases? If not, consider the impact this will have on your walltime and explore ways in which CPU efficiency might be improved for at-scale runs.
The figure below demonstrates scalability for a real analysis conducted on NCI HPC. For each of the 5 runs, the number of total tasks computed was fixed (3,181 X 4 CPU tasks). The CPU efficiency does decline with increasing parallelism, however the line is not too steeply declining (horizontal = perfectly scalable) and efficiency remains above 70%. CPU time remains constant (missing data for nodes=1). We would certainly call this a scalable job.

Other considerations for job and resource optimisation
Our example benchmarking job was a simple one, and your workflow may call for a different approach. You may have an MPI task where the value of interest is the optimal number of nodes rather than optimal number of CPUs, or you may have code that can run on CPU or GPU and require benchmarking to determine optimal walltime vs KSU usage, etc.
Task size
For cases when you can split a large analysis into discrete and independent tasks (like the DNA mapping example) you will need to make decisions about how to size the tasks. Small tasks enable a higher level of parallelism (thus minimising the walltime), however try not to have tasks that require less than a couple of minutes each as this can overload the head node.
For a parallel job on Gadi using the OpenMPI and nci-parallel method (described in the next section), CPUs/nodes are NOT released from the job as they become idle, so if you have many idle CPU while one long-running task completes, your CPU efficiency will be much reduced and your KSU cost increased. This is unlike Artemis job arrays, where each array sub-job consumes an independent slice of CPU resource. So to minimise the impact of idle CPU on parallel jobs, try and size tasks so that they require a similar walltime. For the DNA example, this is achieved by slicing the original input file into smaller files each of 2 millon lines long.
Unequal task walltimes
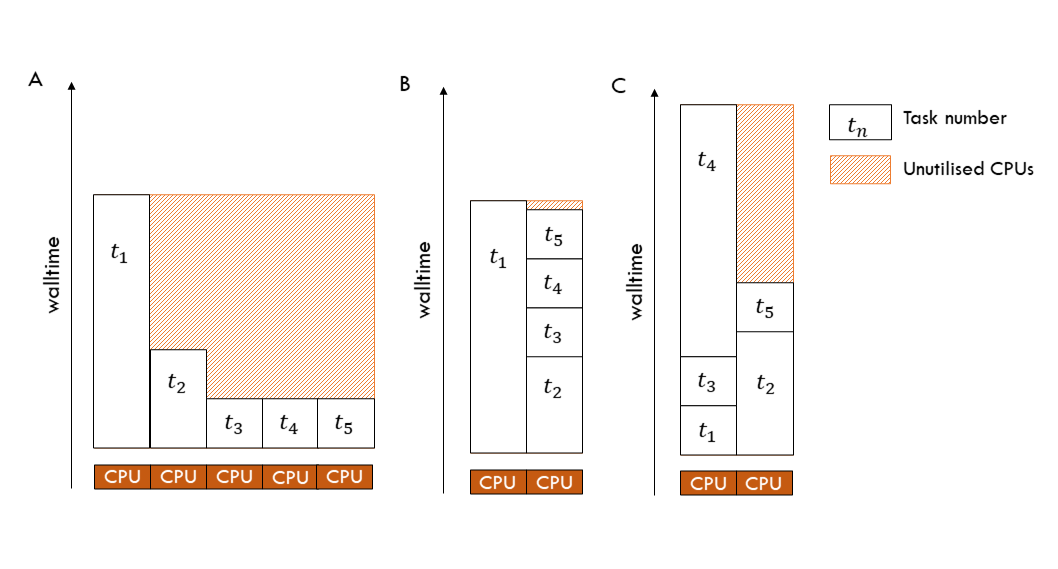
When homogenising the walltime per task is not possible, ordering the tasks by largest to smallest expected walltime, and selecting fewer CPU than required to run all tasks in parallel simultaneously, will enable you to increase CPU efficiency and lower KSU cost. The tasks are assigned to CPU in order, so the smaller tasks can ‘backfill’ CPU as they are freed from executing longer-running tasks.
The figure below demonstrates the impact of task walltime and task order on overall job walltime and idle CPU. Tasks are executed/assigned to CPU in order, t1 to t5. Having an understanding of the expected walltimes for your tasks can help you to minimise idle CPU and thus maximise CPU efficiency and minimise KSU expenditure.
Node type
Gadi has some general-purpose and some specialised nodes, and each has a different charge rate (see Gadi queue limits).
The hardware underlying each queue may be more or less suitable for your job (see Gadi queue structure). Eg the Cascade Lake nodes within the normal and express queues have a faster chip speed than the Cascade Lake hugemem or Broadwell nodes, however only provide 4 GB RAM per CPU. The 256 GB Broadwell nodes offer up to 9 GB RAM per CPU, with a charge rate of only 1.25 SU compared to 3 SU for hugemem.
Performing benchmarking for key jobs in your workflow on a variety of node types can help to optimise efficiency and KSU usage.
One you have these metrics at hand, you are then more equipped to take advantage of lulls on certain queues, for example if the normal queue has some delays however the Broadwell normal queue (normalbw) is underutilised, you can re-shape your job to submit to the queue with available resources. This can help optimise your queue time and give you faster results.
Why parallel jobs are beneficial on Gadi
This and the subsequent section is all about parallel jobs, and there are some very important reason for this:
- While some of you may be thinking it would be very easy to port your Artemis array jobs to Gadi by submitting them with a
forloop, this is actively discouraged on Gadi: the NCI userbase is very large, and the PBS job scheduler and the login nodes would very quickly suffer impaired performance if all users submitted batch jobs in this way. If NCI staff detect a user submitting large quantities of jobs in this way, they will be contacted. - Gadi’s scheduler is designed to favour “short and wide” jobs over “tall and slim” jobs, ie jobs that use more nodes for a shorter walltime will schedule ahead of jobs that use fewer nodes for a longer walltime. Policies behind Gadi’s scheduler are very different to that on Artemis - see ‘Gadi fair share policy’ below.
- Merit-based allocations on Gadi must demonstrate efficient and scalable workflows. Scalability can be defined as parallelisation efficiency - how close the actual speedup is to the ideal speedup given more nodes. Workflows that are not parallel cannot demonstrate scalability.
Gadi fair share policy
Gadi’s ‘fair share’ policy is very different to that of Artemis - there is no ‘fair share weighting’ accruing to your project as you consume CPU hours. The priority of jobs that you submit will decrease compared to the priority of other jobs in the queue if your pro-rata by quarter usage of allocated KSU on Gadi exceeds 50%. For example, if you have consumed 80% of your Q1 KSU within the first month (33% of the quarter), you have consumed more than 50% pro-rata so this will impact your priority in the queue. However, as your job queues for longer, its priority will increase.
In practice, the effect of any lowering of priority by KSU usage on your queue time will hardly/never be noticed by you, as the Gadi queues turn over jobs very quickly.
The key factors that dictate how long your job will queue for is resource availability and other jobs in queue: when will the resources I have requested be available, considering other jobs in the queue? Walltime is important here - of two jobs requesting the same CPUs and RAM in the same queue that are submitted at equal times and with equal priority, one will be assigned to resources first if it has a shorter walltime than the other.
Number of nodes
You may choose to modify the number of nodes you choose for a multi-node or parallel job after assessing the number of jobs in queue (use qstat -q) or free cores (see Gadi live status). When the queues are very busy, resource availability will make it harder to schedule very wide jobs. The maximum number of nodes a job can have on Gadi is 432, however in practice you will achieve better queue times if you split the job into 2 x 216 node jobs or 4 x 108 node jobs, as these are easier to schedule (easier to find 216 or 108 nodes free at once compared to finding 432 free nodes).
Gadi maximum walltimes decrease as the number of CPUs requested increases (see queue limits) so you may need to reduce the number of nodes and sacrifice walltime or split the job into two parallel jobs. We have had success at SIH regularly submitting 8 x 120 node jobs at 30 minutes walltime and having all jobs commence within ten minutes of submission.
In the next section, we will use a resource calculator spreadsheet to demonstrate how you can balance node number and walltime to suit your job requirements.
Questions
![]() Does anyone have any questions about optimisation on NCI Gadi?
Does anyone have any questions about optimisation on NCI Gadi?
Key Points
NCI monitor job performance. Users with consistently underperforming jobs will be singled out!
Time spent benchmarking and optimising key jobs in your workflow can save you considerable walltime and KSU