#Import dask dataframe modules
import dask.dataframe as dd
#import dask
#dask.config.set({"visualization.engine": "cytoscape"})
#NOTE: to run this example (with diagrams) you will need to "pip install graphviz" and donwload graphviz
#https://graphviz.org/download/
import os
#os.environ["PATH"] += os.pathsep + 'C:/APPS/Graphviz/bin'Working with Big Data using Dask
Questions
- Use a modern python library and elegant syntax for performance benefits
- How do I deal with large irregular data and show me some real world examples of Dask?
Objectives
- Intro to Dask concepts and High level datastructures
- Use dask dataframes
- Use dask delayed functions
- Deal with semi-structured and unstructured data in memory efficient and parallel manner
- Show me examples of using Dask on Large Datasets
DASK
Dask is a flexible library for parallel computing in Python.
Dask is composed of two parts: Dynamic task scheduling optimized for computation. This is similar to Airflow, Luigi, Celery, or Make, but optimized for interactive computational workloads. “Big Data” collections like parallel arrays, dataframes, and lists that extend common interfaces like NumPy, Pandas, or Python iterators to larger-than-memory or distributed environments. These parallel collections run on top of dynamic task schedulers.
Dask emphasizes the following virtues:
- Familiar: Provides parallelized NumPy array and Pandas DataFrame objects
- Flexible: Provides a task scheduling interface for more custom workloads and integration with other projects.
- Native: Enables distributed computing in pure Python with access to the PyData stack.
- Fast: Operates with low overhead, low latency, and minimal serialization necessary for fast numerical algorithms
- Scales up: Runs resiliently on clusters with 1000s of cores
- Scales down: Trivial to set up and run on a laptop in a single process
- Responsive: Designed with interactive computing in mind, it provides rapid feedback and diagnostics to aid humans
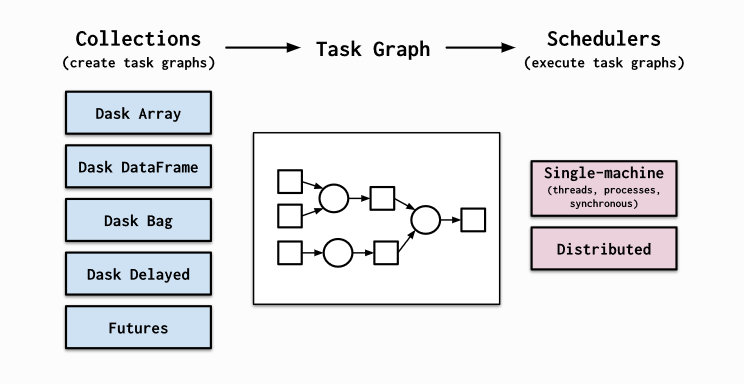
Dask provides high level collections - these are Dask Dataframes, bags, and arrays. On a low level, dask dynamic task schedulers to scale up or down processes, and presents parallel computations by implementing task graphs. It provides an alternative to scaling out tasks instead of threading (IO Bound) and multiprocessing (cpu bound).
A Dask DataFrame is a large parallel DataFrame composed of many smaller Pandas DataFrames, split along the index. These Pandas DataFrames may live on disk for larger-than-memory computing on a single machine, or on many different machines in a cluster. One Dask DataFrame operation triggers many operations on the constituent Pandas DataFrames.
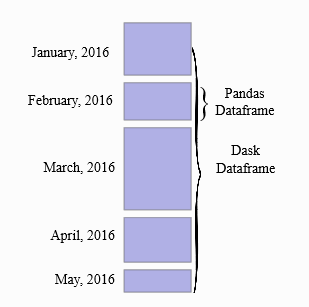
Common Use Cases: Dask DataFrame is used in situations where Pandas is commonly needed, usually when Pandas fails due to data size or computation speed: - Manipulating large datasets, even when those datasets don’t fit in memory - Accelerating long computations by using many cores - Distributed computing on large datasets with standard Pandas operations like groupby, join, and time series computations
Dask Dataframes may not be the best choice if: your data fits comfortable in RAM - Use pandas only! If you need a proper database. You need functions not implemented by dask dataframes - see Dask Delayed.
Dask Dataframes
We will load in some data to explore.
# Setup a parlalle LocalCluster that makes use of all the cores and RAM we have on a single machine
from dask.distributed import Client, LocalCluster
cluster = LocalCluster()
# explicitly connect to the cluster we just created
client = Client(cluster)
client/Users/darya/miniconda3/envs/geopy/lib/python3.9/site-packages/distributed/node.py:183: UserWarning: Port 8787 is already in use.
Perhaps you already have a cluster running?
Hosting the HTTP server on port 54995 instead
warnings.warn(Client
Client-eac15e50-22ac-11ed-9bb4-fe453513c759
| Connection method: Cluster object | Cluster type: distributed.LocalCluster |
| Dashboard: http://127.0.0.1:54995/status |
Cluster Info
LocalCluster
fbc990ef
| Dashboard: http://127.0.0.1:54995/status | Workers: 5 |
| Total threads: 10 | Total memory: 32.00 GiB |
| Status: running | Using processes: True |
Scheduler Info
Scheduler
Scheduler-0247a9eb-a609-475c-b8e5-1e5c3e496e65
| Comm: tcp://127.0.0.1:54998 | Workers: 5 |
| Dashboard: http://127.0.0.1:54995/status | Total threads: 10 |
| Started: Just now | Total memory: 32.00 GiB |
Workers
Worker: 0
| Comm: tcp://127.0.0.1:55037 | Total threads: 2 |
| Dashboard: http://127.0.0.1:55039/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:55002 | |
| Local directory: /var/folders/1b/_jymrbj17cz6t7cxdl86xshh0000gr/T/dask-worker-space/worker-g_vn8opz | |
Worker: 1
| Comm: tcp://127.0.0.1:55044 | Total threads: 2 |
| Dashboard: http://127.0.0.1:55053/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:55004 | |
| Local directory: /var/folders/1b/_jymrbj17cz6t7cxdl86xshh0000gr/T/dask-worker-space/worker-uxx2_0xo | |
Worker: 2
| Comm: tcp://127.0.0.1:55043 | Total threads: 2 |
| Dashboard: http://127.0.0.1:55045/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:55001 | |
| Local directory: /var/folders/1b/_jymrbj17cz6t7cxdl86xshh0000gr/T/dask-worker-space/worker-6v08k6vd | |
Worker: 3
| Comm: tcp://127.0.0.1:55051 | Total threads: 2 |
| Dashboard: http://127.0.0.1:55055/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:55005 | |
| Local directory: /var/folders/1b/_jymrbj17cz6t7cxdl86xshh0000gr/T/dask-worker-space/worker-fd9fzs7b | |
Worker: 4
| Comm: tcp://127.0.0.1:55057 | Total threads: 2 |
| Dashboard: http://127.0.0.1:55058/status | Memory: 6.40 GiB |
| Nanny: tcp://127.0.0.1:55003 | |
| Local directory: /var/folders/1b/_jymrbj17cz6t7cxdl86xshh0000gr/T/dask-worker-space/worker-wsov5c3n | |
All materials copyright Sydney Informatics Hub, University of Sydney